Abstract
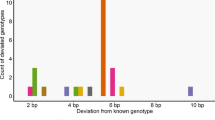
Several methods have been applied to calculate genotyping error rates (GER) for non-invasive population size estimations. However, there is a lack of comparability between these methods. Here we focused on the comparison of methods for determination of GER within one study using faeces samples of wild boars (Sus scrofa). Error rates were calculated by (1) comparison of reference tissue samples and rectum faeces samples (2) the number of deviations between replicates and the assumed consensus genotypes, (3) re-analysis of a subsample interpreted by allelic and genotype comparisons, and (4) a blind-test of anonymously subdivided faecal samples. The error rates differed widely between these four methods (0–57.5 %) and underline the need of a consensus approach. The blind-test resulted in a GER of 4.3 %. We recommend conducting such a blind-test for estimating realistic GER when starting a pilot study in wildlife forensics.


Similar content being viewed by others
References
Arrendal J, Vila C, Bjorklund M (2007) Reliability of noninvasive genetic census of otters compared to field censuses. Conserv Genet 8:1097–1107
Bayes M, Smith K, Alberts S, Altmann J, Bruford M (2000) Testing the reliability of microsatellites typing from feacal DNA. Conserv Genet 1:173–176
Beja-Pereira A, Oliveira R, Alves PC, Schwartz MK, Luikart G (2009) Advancing ecological understandings through technological transformations in noninvasive genetics. Mol Ecol Resour 9(5):1279–1301
Bonin A, Bellemain E, Bronken Eidesen P, Pompanon F, Brochmann C, Taberlet P (2004) How to track and assess genotyping errors in population genetics. Mol Ecol 13(11):3261–3273
Bortz J, Doering N (2002) Forschungsmethoden und evaluation. Springer, Berlin
Broquet T, Petit E (2004) Quantifying genotyping errors in noninvasive population genetics. Mol Ecol 13(11):3601–3608
Broquet T, Menard N, Petit E (2007) Noninvasive population genetics: a review of sample source, diet, fragment length and microsatellite motif effects on amplification success and genotyping error rates. Conserv Genet 8(1):249–260
Creel S, Spong G, Sands JL, Rotella J, Zeigle J, Joe L, Murphy KM, Smith D (2003) Population size estimation in Yellowstone wolves with error-prone noninvasive microsatellite genotypes. Mol Ecol 12(7):2003–2009
Deagle EB, Eveson JP, Jarman SM (2006) Quantification in DNA recovered from highly degraded samples- a case study on DNA in faeces. Front Zool 3:11
Ferrando A, Lecis R, Domingo-Roura X, Ponsa M (2008) Genetic diversity and individual identification of reintroduced otters (Lutra lutra) in north-eastern Spain by DNA genotyping of spraints. Conserv Genet 9(1):129–139
Flagstad O, Roed K, Stacy JE, Jakobsen KS (1999) Reliable noninvasive genotyping based on excremental PCR of nuclear DNA purified with a magnetic bead protocol. Mol Ecol 8(5):879–883
Frantz AC, Pope LC, Carpenter PJ, Roper TJ, Wilson GJ, Delahay RJ, Burke T (2003) Reliable microsatellite genotyping of the Eurasian badger (Meles meles) using faecal DNA. Mol Ecol 12(6):1649–1661
Garnier JN, Bruford MW, Goossens B (2001) Mating system and reproductive skew in the black rhinoceros. Mol Ecol 10(8):2031–2041
Hajkova P, Zemanova B, Roche K, Hajek B (2009) An evaluation of field and noninvasive genetic methods for estimating Eurasian otter population size. Conserv Genet 10(6):1667–1681
Harris RB, Winnie J, Amish SJ, Beja-Pereira A, Godinho R, Costa V, Luikart G (2010) Argali Abundance in the Afghan Pamir using capture-recapture modeling from fecal DNA. J Wildlife Manage 74(4):668–677
Hedmark E, Flagstad Ø, Segerstroem P, Persson J, Landa A, Ellegren H (2004) DNA-based individual and sex identification from wolverine (Gulo gulo) faeces and urine. Conserv Genet 5:405–410
Hoffman JI, Amos W (2005) Microsatellite genotyping errors: detection approaches, common sources and consequences for paternal exclusion. Mol Ecol 14(2):599–612
Idaghdour Y, Broderick D, Korrida A (2003) Faeces as a source of DNA for molecular studies in a threatened population of great bustards. Conserv Genet 4(6):789–792
Kolodziej K, Theissinger K, Brün J, Schulz H.K., Schulz R. (2011) Determination of the minimum number of microsatellite markers for individual genotyping in wild boar (Sus scrofa) using a test with close relatives. Eur J Wildl Res published online, doi: 10.1007/s10344-011-0588-9
Lampa S, Gruber B, Henle K, Hoehn M (2008) An optimisation approach to increase DNA amplification success of otter faeces. Conserv Genet 9(1):201–210
Lucchini V, Fabbri E, Marucco F, Ricci S, Boitani L, Randi E (2002) Noninvasive molecular tracking of colonizing wolf (Canis lupus) packs in the western Italian Alps. Mol Ecol 11(5):857–868
Maudet C, Luikart G, Dubray D, von Hardenberg A, Taberlet P (2004) Low genotyping error rates in wild ungulate faeces sampled in winter. Mol Ecol Notes 4(4):772–775
Murphy M, Kendall K, Robinson A, Waits L (2007) The impact of time and field conditions on brown bear (Ursus arctos) faecal DNA amplification. Conserv Genet 8(5):1219–1224
Paetkau D (2003) An empirical exploration of data quality in DNA-based population inventories. Mol Ecol 12(6):1375–1387
Pompanon F, Bonin A, Bellemain E, Taberlet P (2005) Genotyping errors: causes, consequences and solutions. Nat Rev Genet 6(11):847–859
Prugh L, Ritland C, Arthur S, Krebs C (2005) Monitoring coyote population dynamics by genotyping faeces. Mol Ecol 14(5):1585–1596
Taberlet P, Luikart G (1999) Non-invasive genetic sampling and individual identification. Biol J Linn Soc 68(1–2):41–55
Taberlet P, Griffin S, Goossens B, Questiau S, Manceau V, Escaravage N, Waits LP, Bouvet J (1996) Reliable genotyping of samples with very low DNA quantities using PCR. Nucleic Acids Res 24(16):3189–3194
Valière N (2002) GIMLET: a computer program for analysing genetic individual identification data. Mol Ecol Notes 2(3):377–379
Valière N, Bonenfant C, Toigo C, Luikart G, Gaillard JM, Klein F (2007) Importance of a pilot study for non-invasive genetic sampling: genotyping errors and population size estimation in red deer. Conserv Genet 8(1):69–78
Wehausen JD, Ramey RR 2nd, Epps CW (2004) Experiments in DNA extraction and PCR amplification from bighorn sheep feces: the importance of DNA extraction method. J Hered 95(6):503–509
Wilberg MJ, Dreher BP (2004) Genecap: a programm for analysis of multilocus genotype data for non-invasive sampling and capture-recapture population estimation. Mol Ecol Notes 4(4):783–785
Acknowledgments
We wish to thank D. Huckschlag for good advices for the manuscript. We also thank R. Heydenreich for proofreading of this manuscript. Furthermore, we are grateful to T. Bürgi for technical assistance. This project was supported by the Foundation “Rheinland-Pfalz für Innovation” and the Ministry for Environment, Forestry and Consumer Protection, Rhineland-Palatinate. K.K. was supported through a Ph.D. scholarship from the Lotto Foundation Rhineland-Palatinate.
Author information
Authors and Affiliations
Corresponding authors
Rights and permissions
About this article
Cite this article
Kolodziej, K., Schulz, H.K., Theissinger, K. et al. Comparison of established methods for quantifying genotyping error rates in wildlife forensics. Conservation Genet Resour 5, 287–292 (2013). https://doi.org/10.1007/s12686-012-9729-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12686-012-9729-z