Abstract
Background
Spi-1 proto-oncogene (SPI1), which encodes an ETS-domain transcription factor, can activate gene expression in myeloid and lymphoid lineages. The role of SPI1 in the tumor immune microenvironment in clear cell renal cell carcinoma (ccRCC) remains unknown. In this study, we investigated the possible role of SPI1 in ccRCC using an independent cohort and a comprehensive bioinformatics analysis.
Materials and methods
Quantitative real-time PCR, western blot and immunohistochemistry assays were used to compare the SPI1 expression levels between ccRCC tissues and normal tissues, analyze the relationships between SPI1 and CD68, CD8, CD4 expression levels, and explore the link between SPI1 and the efficacy of immunotherapy in our cohort. Tumor Immune Estimation Resource, UALCAN, cBioPortal, TISIDB database, and LinkedOmics database were used in our study.
Results
SPI1 expression level was higher in ccRCC bulk tissues than in normal bulk tissues. SPI1 was an independent prognostic factor for poor overall survival and progression-free survival in patients with ccRCC. SPI1 expression was strongly related to the infiltration of immune cells and immune-related molecules. SPI1 was more highly expressed in tumor-infiltrating immune cells rather than in cancer cells. Non-responders to immunotherapy against ccRCC were more likely to express higher SPI1 levels than responders. Genes co-expressed with SPI1 primarily correlated with immune-related pathways.
Conclusions
SPI1 expression in tumor bulk tissues is associated with disease progression and poor prognosis, as well as high expression levels of immune markers and infiltration of immune cells. SPI1 can be used as a prognostic biomarker to monitor and evaluate immunotherapy efficacy.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
Clear cell renal cell carcinoma (ccRCC), a tumor with high immune infiltration, is a major histological subtype of renal cell carcinoma [1]. Although cancer treatment, particularly cancer immunotherapy, has greatly advanced in recent years, patients with metastatic and advanced ccRCC have poor prognoses [2]. Additionally, effective markers for monitoring and evaluating immunotherapy efficacy are lacking.
The tumor immune microenvironment (TME), which contains tumor cells, immune cells, their surrounding stroma, and extracellular components, affects tumor progression and immunotherapy efficacy [3]. Tumor progression or suppression is most dependent on the immunosuppressive or immunostimulatory state [4]. T-cell exhaustion and tumor-infiltrating lymphocytes (TILs) are associated with an exhausted phenotype and limited anti-tumor activity [5]. Tumor-associated macrophages, myeloid-derived suppressor cells (MDSCs), Treg cells, and neutrophils are known to promote tumor progression by creating an immunosuppressive tumor environment. Modulated by cytokines and chemokines, tumor-infiltrating immune cells (TIICs) interact with cancer cells in an extensive and dynamic manner and play essential roles in TME [6]. TIICs not only affect tumor development, progression, and response to immunotherapy, but also predict clinical outcomes in patients with cancer [6]. Although several molecules (PD-1, CTLA4, LAG3, TIM3, and TIGIT) in immune cells may be useful markers for prognosis and immunotherapy efficacy in cancer [7], more reliable and effective biomarkers for individualized prognosis and therapy selection are urgently needed.
Spi-1 proto-oncogene (SPI1) encodes PU.1 (a member of the E26-transformation-specific transcription factor family), an essential factor in signal transmission in the immune system and the development of myeloid cells and lymphocytes [8]. Macrophage differentiation is regulated by SPI1-related gene sets [9]. In addition, SPI1 is important in malignant diseases. SPI1 has been identified to facilitate glycolytic processes and promote colon cancer progression [10]. The SPI1-METTL14-MYB/MYC signaling axis participates in myelopoiesis and leukemogenesis [11]. Transcriptional activation of SPI1 can enhance the tumorigenic potential of cervical cancer cells through its effect on PARP9 [12]. Importantly, Li et al. demonstrated that the transcription factor SPI1 can promote the expression of CD86, CCL4/CXCL10/CX3CL1, and MHC-II, thereby increasing immune infiltration [13]. However, the relationship between of SPI1 expression and immune cell infiltration in ccRCC remains unclear.
In our cohort, SPI1 was significantly overexpressed in ccRCC stroma and was closely related to several clinicopathological parameters of ccRCC, including the Furman grade, T stage, nodal metastasis, and metastasis. In ccRCC bulk tissues, DNA methylation levels of SPI1 was lower than that in normal bulk tissues. Furthermore, SPI1 was found to be an independent indicator of poor OS and PFS in patients with ccRCC. Interestingly, immunohistochemistry revealed that SPI1 was more highly expressed in TIICs rather than in cancer cells, consistent with the SPI1 expression in renal cell lines. SPI1 expression and methylation status were strongly correlated with immune cell infiltration and immune-related molecules in ccRCC. For immunotherapy in RCC, non-responders were more likely to express higher SPI1 levels than responders. Genes co-expressed with SPI1 are associated with various immune processes. Thus, SPI1 expression level may be a marker of poor prognosis and immunotherapy efficacy associated with immune cell infiltration.
2 Materials and methods
2.1 Patients and samples
A ccRCC tissue microarray (TMA) containing primary tumor tissues and paired adjacent normal tissues of 183 patients was obtained from the Department of Urology, The First Medical Center, Chinese PLA General Hospital, Beijing, China. Primary tumor tissues from seven patients undergoing immunotherapy plus targeted therapy (pembrolizumab plus axitinib) after radical nephrectomy were also paraffin-embedded and sectioned for subsequent experiments. This study was approved by the Ethics Committee of the Chinese PLA General Hospital.
2.2 Clinical cohorts
Patient data from three prospective clinical trials of the anti-PD-1 antibody nivolumab in ccRCC, including CheckMate 009 (CM-009; NCT01358721) [14], CheckMate 010 (CM-010; NCT01354431) [15], and CheckMate 025 (CM-025; NCT01668784) [16] were used to explore the relationship between SPI1 expression and immunotherapy efficacy in this study [17]. We obtained 129 ccRCC patients with primary tumor RNA sequencing data and complete clinical efficacy of nivolumab (Additional file 3: Table S3). Clinical benefit (CB) encompassed patients who achieved complete responses or partial responses, or stable disease with tumor shrinkage and PFS of at least 6 months. No clinical benefit (NCB) encompassed patients who achieved progressive disease and PFS less than 3 months. All other patients were classified as having intermediate clinical benefit (ICB) [17]. Patients were stratified into low or high groups based on SPI1 expression, using the median RNA expression of SPI1 as the cut-off value. Correlations between SPI1 expression and clinical efficacy of nivolumab was analyzed by the Pearson’s Chi squared test.
2.3 Quantitative real-time PCR (qRT-PCR)
qRT-PCR was used to determine SPI1 mRNA expression levels in ccRCC and normal bulk tissues. To extract total RNA, an RNA-Quick Purification Kit (RN001; ESscience Biotech, China) was used. Complementary DNA was synthesized using a kit from BIO-RAD, iScriptTM cDNA Synthesis Kit (1708890). For quantitative PCR, iTaq Universal SYBR® Green Supermix (1725120, Bio-Rad, USA) was used. The 2-∆∆Ct method was used to analyze the relative mRNA expression level of SPI1 normalized to the level of peptidylprolyl isomerase A (PPIA). The forward primer sequence for SPI1: GTGCCCTATGACACGGATCTA, reverse primer: AGTCCCAGTAATGGTCGCTAT. The process was performed with gene-specific primers and SYBR Green detection using QuantStudio 3 (Applied Biosystems).
2.4 Western blot
Western blot assays were performed in accordance with standard techniques as previously reported [18]. Antibodies against SPI1 (66618–2-lg; Proteintech) andβ-Tubulin (BE0025; EASYBIO) were used.
2.5 Immunohistochemistry (IHC)
IHC staining was used to examine the expression level of SPI1 protein in cancerous and paracancerous tissues from our cohort. IHC staining of SPI1 (Atlas, HPA055653), CD68 (Abcam, ab955), CD8 (Proteintech, 66868–1-Ig), and CD4 (Abcam, ab133616) was performed on TMA tissues. We followed standard protocols, as described previously [19]. The slides were scaned using a Panamic 250 Flash III slide scanner (3DHISTECH Ltd, Budapest, Hungary) and images were analyzed using CaseViewer software (3DHISTECH Ltd, Budapest, Hungary) [20]. Positive cells were counted in five random fields (400X) [21].
2.6 Cell culture
Fetal bovine serum (FBS) and different type of media including high glucose-DMEM, MEM, RPMI-1640, and McCoy’s 5A was purchased from EVERY GREEN (Hangzhou, China) and VIVICUM bioscience (Beijing, China), respectively. The human embryonic kidney derived cell line HEK293T, human renal tubular epithelial cell line HKC and HK2, human ccRCC cell lines SN12, A498, 786O, ACHN, OS-RC-2, and Caki-1 were originally purchased from American Type Culture Collection. The murine renal carcinoma cell line RENCA were purchased from Peking Union Medical College. RAW246.7 macrophages were purchased from BeNa Culture Collection (Beijing, China). Cell culture conditions were as describe [22].
2.7 Tumor immune estimation resource (TIMER) analysis
In the TIMER (https://cistrome.shinyapps.io/timer/) database [23], correlation modules and gene modules were used to analyze the relationship between SPI1 expression and gene markers of immune cells [24, 25].
2.8 UALCAN analysis
Using the UALCAN (http://ualcan.path.uab.edu/) database [26, 27], the expression and methylation status of SPI1 were examined in different sample types of ccRCC by Wilcoxon rank sum test.
2.9 TISIDB database analysis
Using the TISIDB database (http://cis.hku.hk/TISIDB) [28], the associations between the methylation status of SPI1 and infiltrations of lymphocytes, immunomodulators, chemokines, and chemokine receptors were determined.
2.10 cBioPortal analysis
Using the cBioPortal (www.cbioportal.org) database [29], the relationship between the mRNA expression and methylation levels of SPI1 were determined by Spearman correlation coefficient analysis. (Additional file 4: Fig. S1b).
2.11 LinkedOmics database analysis
In the LinkedOmics (http://www.linkedomics.org/login.php) database [30], differentially expressed genes related to SPI1 in ccRCC were identified. Correlation analysis using the Pearson correlation coefficient is shown in Additional file 2: Table S2.
2.12 Statistical analysis
Histogram generation was performed using GraphPad Prism software (version 8.0). Scatter plots were generated using the “ggplot” (version 3.3.3) packages in R. “survival” (version 3.2–10) package and “survminer” (version 0.4.9) package in R were used to draw survival curves. Wilcoxon rank sum test was used to analyze the differential expression levels of SPI1 protein between the ccRCC tissues and the adjacent normal tissues in our cohort. Correlations between SPI1 expression and clinicopathological parameters were analyzed by the Pearson’s Chi squared test. Patients were stratified into low or high groups based on SPI1 expression, using the median numbers of SPI1 positive cells per 20000um2 as the cut-off value. Univariate and multivariable Cox proportional hazards regression analyses were performed to identify potential prognostic factors associated with OS and PFS using the “survival” package of R. Nomogram analysis was conducted using the “survival” (version 3.2–10) and “rms” (version 6.2–0) packages to establish the risk prediction model. The correlation between SPI1 expression and CD68, CD8, and CD4 expression in ccRCC was analyzed by Spearman correlation coefficient analysis. All statistical analyses were performed using GraphPad Prism version 8.0 software (GraphPad, Inc., La Jolla, CA, USA) and R, version 3.6.3 software (The R Group for Statistical Computing, Vienna, Austria).
3 Results
3.1 SPI1 expression in ccRCC
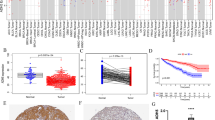
UALCAN database analysis revealed that SPI1 protein and mRNA expression levels were elevated in ccRCC bulk tissues compared to that in normal bulk tissues (Fig. 1a, b). Western blot assay, qRT-PCR analysis and IHC staining confirmed that SPI1 mRNA and protein expression levels were upregulated in ccRCC bulk tissues compared to those in normal bulk tissues (Fig. 1c–e). SPI1 staining was mainly observed in TIICs rather than in tumor cells (Fig. 1e). We evaluated the methylation levels of SPI1 in the ccRCC dataset and found that tumor bulk tissues had significantly lower DNA methylation levels than the normal bulk tissues (Additional file 4: Fig. S1a). Correlation analysis showed that SPI1 mRNA expression was negatively correlated with methylation status (Additional file 4: Fig. S1b). Furthermore, hypomethylation of SPI1 was more likely to occur in late-stage, high-grade tumors, and those with N1 nodal metastasis status (Additional file 4: Fig. S1c–e). Based on these results, SPI1 mRNA and protein levels were higher in ccRCC bulk tissues compared to normal bulk tissues.
SPI1 expression in ccRCC. a, b SPI1 protein and mRNA expression levels were elevated in ccRCC bulk tissues compared to normal bulk tissues in the UALCAN database. c SPI1protein expression levels in six pairs ccRCC tumor bulk tissues and adjacent normal bulk tissues as well as ccRCC cell lines. d SPI1 mRNA expression levels in eight ccRCC tumor bulk tissues and adjacent normal bulk tissues (e) SPI1 protein expression levels was elevated in ccRCC stroma compared to normal stroma in our cohort. ccRCC, clear cell renal cell carcinoma; *, p < 0.05; **, p < 0.01; ***, p < 0.001
3.2 Prognostic value of SPI1 and its correlation with clinicopathological parameters in ccRCC
The relationships between SPI1 and the clinicopathological features of patients with ccRCC were explored in our cohort with 183 ccRCC patients from the Department of Urology, The First Medical Center, Chinese PLA General Hospital, Beijing, China, and clinicopathological parameters correlated with SPI1 expression, such as Furman grade, T stage, N stage, and M stage were identified (Fig. 2a).High SPI1 expression level was associated with high Furman grade, T stage, positive nodal status, and metastatic status (Fig. 2b–e). SPI1 expression was not associated with number of lymph nodes (regional lymph node metastases involving one node vs. regional lymph node metastases involving more than one node) and the site of metastasis. (data no shown). High SPI1 expression levels predicted poor OS (hazard ratio [HR] = 6.49, p < 0.001) and PFS (hazard ratio [HR] = 7.63, p < 0.001) in patients with ccRCC (Fig. 3a, b). Multivariate Cox regression analysis indicated that SPI1 expression, age, Furman grade, T stage, and M stage were independent prognostic factors for OS in patients with ccRCC (Table 1). SPI1 expression, age, Furman grade, and T stage were independent prognostic factors for PFS in ccRCC patients (Table 2). Finally, the overall five-year survival rate and progression-free five-year survival rate were estimated using nomograms (Fig. 3c, d). Calibration plots of the nomograms for the prediction of five-year OS (Fig. 3e) and PFS (Fig. 3f) are shown. These results suggest that SPI1 is associated with a poor clinical prognosis in ccRCC.
Relationship between SPI1 expression and clinicopathological parameters in ccRCC. a Clinicopathological parameters correlated with SPI1 expression. b–e High SPI1 expression level was associated with high Furman grade (b), T stage (c), positive nodal status (d), and metastasis status (e). ccRCC, clear cell renal cell carcinoma; *, p < 0.05; **, p < 0.01; ***, p < 0.001
Prognostic value of SPI1. a, b High SPI1 expression level predicted poor OS (a) and PFS b in ccRCC. c, d OS (c) and PFS (d) predictions of patients with ccRCC at five years after surgery are shown in nomograms. e, f Calibration plots of the nomograms for the prediction of five-year OS e and PFS f. ccRCC clear cell renal cell carcinoma; OS overall survival; PFS progression-free survival
3.3 SPI1 is widely expressed in immune cell types correlating with immune infiltration and immunotherapy efficacy in ccRCC
Tumor tissues contain large numbers of non-tumor cells, of which immune cells represent a major fraction [31]. According to the results of IHC staining, SPI1 was mainly expressed in the cell nuclei of immune cells, rather than in tumor cells, implying that SPI may be a biomarker of immune cells infiltration in TME. We evaluated the correlations between SPI1 expression and CD68, CD8, and CD4 expression in ccRCC using our cohort and found that SPI1 expression level was positively correlated with infiltrating levels of CD68 + macrophages, CD4 + T cells, and CD8 + T cells (Fig. 4a). The TME with high SPI1 expression level can simultaneously maintain high macrophage, CD8 + T cell, and CD4 + T cell infiltration (Fig. 4b). To further explore the relationship between SPI1 and immunity in ccRCC, we performed a correlation analysis adjusted for tumor purity between SPI1 and gene markers of immune cells and stromal cells using the TIMER platform. SPI1 was strongly correlated with gene markers of CD8 + T cells, Tregs, monocytes, tumor-associated macrophages, M2 macrophages, neutrophils, dendritic cells, regulatory T cells (Tregs), and T cell exhaustion (Table 3). Thus, SPI1 can be a biomarker of immune infiltration in the TME of ccRCC. To investigate the association between SPI1 expression and immunotherapy efficacy in RCC, IHC staining of SPI1 was performed in six patients with RCC who had received at least four cycles of pembrolizumab plus axitinib at our center (Fig. 5a, b). SPI1 expression levels were higher in non-responders (progressive disease or stable disease) than responders (complete disease or partial disease). We obtained 129 ccRCC patients data from three prospective clinical trials of the anti-PD-1 antibody nivolumab[14; 15; 16]. We found that patients with lower SPI1 expression were more likely to had clinical benefit Additional file 4: Fig. S3). Therefore, SPI1 may be a marker of immune cells associated with immune infiltration and immunotherapy efficacy in ccRCC.
Relationship between SPI1 expression and CD68 + macrophages, CD4 + T cells, and CD8 + T cells infiltration. a SPI1 expression correlated positively with CD68 + macrophages, CD4 + T cells, and CD8 + T cells infiltration in our cohort. b SPI1, CD68 + , CD8 + , and CD4 + expression levels in the same patients are shown by IHC. ccRCC, clear cell renal cell carcinoma; *, p < 0.05; **, p < 0.01; ***, p < 0.001; IHC, immunohistochemistry
Relationship between SPI1 expression and immunotherapy efficacy. a SPI1 expression in three responders receiving immunotherapy. b SPI1 expression in four non-responders receiving immunotherapy. Responders included patients with complete disease and partial disease, and non-responders included patients with progressive disease and stable disease
3.4 Methylation status of SPI1 associated with immune infiltrates is a novel prognostic marker in ccRCC
Unlike the positive relationship between the expression of SPI1 and lymphocytes, hypermethylation of SPI1 was negatively correlated with lymphocytes in the TISIDB database (Additional file 2: Fig. S2a). Similarly, the methylation status of SPI1 correlated negatively with both immunoinhibitors, such as LGALS9, PDCD1, LAG3, and TIGIT, and immunostimulators, such as CD86, CD48, TNFS13B, and TNFRSF8, (Additional file 4: Fig. S2b, c). In addition, a negative relationship was observed between the methylation status of SPI1 and chemokines and receptors, such as CXCL16, CCL5, CXCR6, and CCR1 (Additional file 4: Fig. S2d). DNA methylation can be a prognostic biomarker for RCC survival [32,33,34]. The correlation between the methylation status of SPI1 and survival at CpG sites targeted for SPI1 in ccRCC was explored in MethSurv. The results showed that four CpG sites, including cg03106245, cg06147863, cg07698783, and cg15982099 in SPI1, could be prognostic markers for OS in ccRCC (Additional file 1: Table S1). Therefore, the methylation level of SPI1 is associated with immune infiltrates and may be a novel prognostic marker for survival in ccRCC.
3.5 SPI1 co-expression network in ccRCC
To predict the biological function of SPI1, we performed gene co-expression network analysis of SPI1 using the LinkedOmics database [30]. We identified 8,264 and 5,265 positively and negatively correlated genes, respectively (p < 0.05) (Additional file 2: Table S2). The top 50 genes that were positively and negatively correlated with SPI1 expression are shown in the heat maps, respectively (Fig. 6a, b). To clarify the biological functions of genes co-expressed with SPI1, we conducted Gene Ontology enrichment analysis using Gene Set Enrichment Analysis. We found that these genes were linked to mast cell activation, adaptive immune response, T cell activation, immune response-regulating signaling pathways, and other immune processes (Fig. 6c). We also performed Kyoto Encyclopaedia of Genes and Genomes pathway analysis to evaluate the functional enrichment of the co-expressed SPI1 genes and observed enrichment in various immune processes (Fig. 6d). Thus, the SPI1 expression network is associated with immune and inflammatory responses in ccRCC.
Genes coexpressed with SPI1 in KIRC using the LinkedOmics database. a, b The top 50 genes positively and negatively associated with SPI1 shown in the heat maps. Blue represents negatively linked genes and red represents positively linked genes. c, d Gene Ontology annotations c and Kyoto Encyclopedia of Genes and Genomes pathways (d) of SPI1 in KIRC cohort. KIRC, kidney renal clear cell carcinoma
4 Discussion
SPI1 dysregulation has been reported in many cancers [10]. SPI1 can be a prognostic marker and immunotherapeutic target for gastric cancer patients [35]. Li et al. found that SPI1 is associated with immune infiltration during oncogenesis [13]. However, the role of SPI1 in ccRCC is unknown, and the relationship between SPI1 and immune infiltrates in ccRCC remains unclear. Thus, we comprehensively examined the clinical role of SPI1 and relationship between SPI1 and immune infiltrates in ccRCC using our cohort and open-access databases.
In this study, high SPI1 expression level predicted poor OS and PFS in ccRCC patients (Tables 1 and 2). There are several explanations for the poor prognosis of patients with high SPI1 expression level. First, SPI1 plays a crucial role in myeloid cells and T cells development. A strong positive relationship between SPI1 expression and macrophages, CD8 + T cells, and CD4 + T cells infiltration was validated via correlation analysis in 183 ccRCC patients. Indeed, macrophages and CD4 + T cells infiltration correlates with poor prognosis in RCC, and our results are consistent with these findings. However, CD8 + T cells infiltration in RCC did not affect clinical outcomes (data not shown). Second, TILs are dominated by M2 macrophages, which represent a major fraction of macrophages in the tumor-associated microenvironment [36]. The TME with high SPI1 expression level can simultaneously maintain high CD68 + macrophages, implying that SPI1 may be located in M2 macrophages and contributes to tumor-associated macrophage-mediated cancer progression (Fig. 3 and Table 3). SPI1 expression was positively and strongly associated with T cell exhaustion markers (LAG3 and PDCD1,), and Treg cell markers (FOXP3 and CCR8) (Table 3), suggesting that high SPI1 expression level inhibits anti-tumor immunity. Third, SPI1 expression was strongly associated with dendritic cell markers (HLA-DPB1, HLA-DRA, HLA-DPA1, and CD11c) and neutrophils (CD11b) (Table 3). The infiltration of dendritic cells and neutrophils in tumors leads to angiogenesis and cancer progression. In summary, high expression level of SPI1 correlates with immune cells infiltration, resulting in an immunosuppressive microenvironment and exhaustion of tumor-specific T cells, leading to poor prognosis.
Although cancer immunotherapy has greatly advanced in recent years, the clinical efficacy of immunotherapy is limited in most patients owning to the immunosuppressive tumor environment [37]. He et al. found that SPI1 is one of the core genes associated with immunotherapy efficacy in triple-negative breast cancers[38]. We found that patients with higher SPI1 expression levels who were treated by immunotherapy had minor clinical benefits (Fig. 5a, b, Additional file 4: Fig. S3). SPI1 was associated with monocyte /macrophage, T cell, neutrophil, and myeloid dendritic cell infiltration (Fig. 4 and Table 3). Monocytes /macrophages, tumor-infiltrating Tregs, and MDSCs that accumulates in TAM weaken the therapeutic efficacy of immunotherapy [39,40,41,42]. This may explain why high SPI1 expression level is associated with poor immunotherapy efficacy.
Genes in the same pathway or biological process tend to be coexpressed. Co expression and functional enrichment analyses suggested that SPI1 expression was positively associated with T cell activation and positive regulation of cell activation (Fig. 6c). This may seem contradictory given that SPI1 expression is prognostic factor for poor survival. This may be because once inflammation persists and T cell activation continues, T cells show loss of interleukin-2 production, cytokine polyfunctionality, and exhausted proliferative capacity [43, 44]. Furthermore, T cell exhaustion is accompanied by increased expression levels of inhibitory receptors (PDCD1, LAG3, CD244, CD160, and TIGIT) [44]. Thus, the sustained activation of T cells in the TME of ccRCC with high SPI1 expression level ultimately leads to an immunosuppressive state, which requires further investigation. In addition to T cell activation and positive regulation of cell activation, coexpressed genes of SPI1 participate in various immune cell responses, suggesting that SPI1 may be associated with various immune-related processes. Recent studies have revealed the potential cause of this association between SPI1 expression, immune infiltration, and immune molecules. Localized to the nucleus of immune cells, the SPI1 transcription factor plays a crucial role in myeloid and T cells development. Thus, high SPI1 expression level correlates with high infiltration levels of macrophages, CD8 + T cells, and CD4 + T cells. Chemokines such as CCL5, CCL4, CXCL9, and CXCL10 secreted by macrophages can be implicated in the recruitment of CD8 + T cells and can promote immune escape by upregulating immunosuppressive molecules (CTLA4, PD1, and LAG3,) (Table 3). Furthermore, cancer cells can secret immune molecules that are dependent on macrophage-derived chemokines to recruit immune cells into TME [45, 46]. In summary, macrophages and T cells with high SPI1 expression levels may interact with cancer cells by secreting chemokines and immune molecules.
Our study has some limitations. First, our cohort included a relatively small number of patients who received immunotherapy. Second, basic experiments are required to validate the biological role of SPI1 and to explore its mechanism in ccRCC immunity.
5 Conclusions
SPI1 is a promising prognostic biomarker for ccRCC associated with poor efficacy of immunotherapy, along with high infiltration of immune cells within the TME.
Data availability
The datasets analyzed for this study can be found in the TIMER database (https://cistrome.shinyapps.io/timer/), the UALCAN database (http://ualcan.path.uab.edu/), the TISIDB database (http://cis.hku.hk/TISIDB), the cBioPortal database (www.cbioportal.org), and the LinkedOmics database (http://www.linkedomics.org/login.php). The raw data from our central supporting the conclusions of this article will be made available by the authors, without undue reservation.
References
Siegel RL, Miller KD, Jemal A. Cancer statistics 2019. CA Cancer J Clin. 2019;69:7–34.
Choueiri TK, Halabi S, Sanford BL, Hahn O, Michaelson MD, Walsh MK, Feldman DR, Olencki T, Picus J, Small EJ, Dakhil S, George DJ, Morris MJ. Cabozantinib versus sunitinib as initial targeted therapy for patients with metastatic renal cell carcinoma of poor or intermediate risk: the alliance A031203 CABOSUN Trial. J Clin Oncol. 2017;35:591–7.
Roma-Rodrigues C, Mendes R, Baptista PV, Fernandes AR. Targeting tumor microenvironment for cancer therapy. Int J Mol Sci. 2019;20:840.
Li L, Li Y, Yang CH, Radford DC, Wang J, Janát-Amsbury M, Kopeček J, Yang J. Inhibition of immunosuppressive tumors by polymer-assisted inductions of immunogenic cell death and multivalent PD-L1 crosslinking. Adv Funct Mater. 2020;30:1908961.
Ma X, Bi E, Lu Y, Su P, Huang C, Liu L, Wang Q, Yang M, Kalady MF, Qian J, Zhang A, Gupte AA, Hamilton DJ, Zheng C, Yi Q. Cholesterol induces CD8(+) T cell exhaustion in the tumor microenvironment. Cell Metab. 2019;30:143-156.e5.
Grivennikov SI, Greten FR, Karin M. Immunity, inflammation, and cancer. Cell. 2010;140:883–99.
Waldman AD, Fritz JM, Lenardo MJ. A guide to cancer immunotherapy: from T cell basic science to clinical practice. Nat Rev Immunol. 2020;20:651–68.
DeKoter RP, Singh H. Regulation of B lymphocyte and macrophage development by graded expression of PU.1. Science. 2000;288:1439–41.
Weigelt K, Lichtinger M, Rehli M, Langmann T. Transcriptomic profiling identifies a PU.1 regulatory network in macrophages. Biochem Biophys Res Commun. 2009;380:308–12.
Wang J, Wang X, Guo Y, Ye L, Li D, Hu A, Cai S, Yuan B, Jin S, Zhou Y, Li Q, Zheng L, Tong Q. Therapeutic targeting of SPIB/SPI1-facilitated interplay of cancer cells and neutrophils inhibits aerobic glycolysis and cancer progression. Clin Transl Med. 2021;11:e588.
Weng H, Huang H, Wu H, Qin X, Zhao BS, Dong L, Shi H, Skibbe J, Shen C, Hu C, Sheng Y, Wang Y, Wunderlich M, Zhang B, Dore LC, Su R, Deng X, Ferchen K, Li C, Sun M, Lu Z, Jiang X, Marcucci G, Mulloy JC, Yang J, Qian Z, Wei M, He C, Chen J. METTL14 inhibits hematopoietic stem/progenitor differentiation and promotes leukemogenesis via mRNA m(6)A modification. Cell Stem Cell. 2018;22:191-205.e9.
Tao L, Wang X, Zhou Q. Long noncoding RNA SNHG16 promotes the tumorigenicity of cervical cancer cells by recruiting transcriptional factor SPI1 to upregulate PARP9. Cell Biol Int. 2020;44:773–84.
Li J, Shi H, Yuan Z, Wu Z, Li H, Liu Y, Lu M, Lu M. The role of SPI1-TYROBP-FCER1G network in oncogenesis and prognosis of osteosarcoma, and its association with immune infiltration. BMC Cancer. 2022;22:108.
Choueiri TK, Fishman MN, Escudier B, McDermott DF, Drake CG, Kluger H, Stadler WM, Perez-Gracia JL, McNeel DG, Curti B, Harrison MR, Plimack ER, Appleman L, Fong L, Albiges L, Cohen L, Young TC, Chasalow SD, Ross-Macdonald P, Srivastava S, Jure-Kunkel M, Kurland JF, Simon JS, Sznol M. Immunomodulatory activity of nivolumab in metastatic renal cell carcinoma. Clinical cancer research : an official journal of the American Association for Cancer Research. 2016;22:5461–71.
Motzer RJ, Rini BI, McDermott DF, Redman BG, Kuzel TM, Harrison MR, Vaishampayan UN, Drabkin HA, George S, Logan TF, Margolin KA, Plimack ER, Lambert AM, Waxman IM, Hammers HJ. Nivolumab for metastatic renal cell carcinoma: results of a randomized phase II trial. J Clin Oncol. 2015;33:1430–7.
Motzer RJ, Tannir NM, McDermott DF, Arén Frontera O, Melichar B, Choueiri TK, Plimack ER, Barthélémy P, Porta C, George S, Powles T, Donskov F, Neiman V, Kollmannsberger CK, Salman P, Gurney H, Hawkins R, Ravaud A, Grimm MO, Bracarda S, Barrios CH, Tomita Y, Castellano D, Rini BI, Chen AC, Mekan S, McHenry MB, Wind-Rotolo M, Doan J, Sharma P, Hammers HJ, Escudier B. Nivolumab plus Ipilimumab versus Sunitinib in advanced renal-cell carcinoma. New Eng J Med. 2018;378:1277–90.
Braun DA, Hou Y, Bakouny Z, Ficial M, Sant’ Angelo M, Forman J, Ross-Macdonald P, Berger AC, Jegede OA, Elagina L, Steinharter J, Sun M, Wind-Rotolo M, Pignon JC, Cherniack AD, Lichtenstein L, Neuberg D, Catalano P, Freeman GJ, Sharpe AH, McDermott DF, Van Allen EM, Signoretti S, Wu CJ, Shukla SA, Choueiri TK. Interplay of somatic alterations and immune infiltration modulates response to PD-1 blockade in advanced clear cell renal cell carcinoma. Nature Med. 2020;26:909–18.
Wang H, Fan Y, Chen W, Lv Z, Wu S, Xuan Y, Wang C, Lu Y, Guo T, Shen D, Zhang F, Huang Q, Gao Y, Li H, Ma X, Wang B, Huang Y, Zhang X. Loss of CMTM6 promotes DNA damage-induced cellular senescence and antitumor immunity. Oncoimmunology. 2022;11:2011673.
Zhao T, Ren H, Li J, Chen J, Zhang H, Xin W, Sun Y, Sun L, Yang Y, Sun J, Wang X, Gao S, Huang C, Zhang H, Yang S, Hao J. LASP1 is a HIF1α target gene critical for metastasis of pancreatic cancer. Can Res. 2015;75:111–9.
Arroyo-Crespo JJ, Armiñán A, Charbonnier D, Deladriere C, Palomino-Schätzlein M, Lamas-Domingo R, Forteza J, Pineda-Lucena A, Vicent MJ. Characterization of triple-negative breast cancer preclinical models provides functional evidence of metastatic progression. Int J Cancer. 2019;145:2267–81.
Li J, Chen Y, Wan J, Liu X, Yu C, Li W. ABT-263 enhances sorafenib-induced apoptosis associated with Akt activity and the expression of Bax and p21((CIP1/WAF1)) in human cancer cells. Br J Pharmacol. 2014;171:3182–95.
Lv Z, Cui B, Huang X, Feng HY, Wang T, Wang HF, Xuan YD, Li HZ, Ma X, Huang Y, Zhang X. FGL1 as a novel mediator and biomarker of malignant progression in clear cell renal cell carcinoma. Front Oncol. 2021;11:756843.
Li T, Fan J, Wang B, Traugh N, Chen Q, Liu JS, Li B, Liu XS. TIMER: a web server for comprehensive analysis of tumor-infiltrating immune cells. Can Res. 2017;77:e108–10.
Li B, Severson E, Pignon JC, Zhao H, Li T, Novak J, Jiang P, Shen H, Aster JC, Rodig S, Signoretti S, Liu JS, Liu XS. Comprehensive analyses of tumor immunity: implications for cancer immunotherapy. Genome Biol. 2016;17:174.
Li B, Senbabaoglu Y, Peng W, Yang ML, Xu J, Li JZ. Genomic estimates of aneuploid content in glioblastoma multiforme and improved classification. Clin Cancer Res. 2012;18:5595–605.
Chandrashekar DS, Karthikeyan SK, Korla PK, Patel H, Shovon AR, Athar M, Netto GJ, Qin ZS, Kumar S, Manne U, Creighton CJ, Varambally S. UALCAN: an update to the integrated cancer data analysis platform. Neoplasia. 2022;25:18–27.
Chandrashekar DS, Bashel B, Balasubramanya SAH, Creighton CJ, Ponce-Rodriguez I, Chakravarthi B, Varambally S. UALCAN: a portal for facilitating tumor subgroup gene expression and survival analyses. Neoplasia. 2017;19:649–58.
Ru B, Wong CN, Tong Y, Zhong JY, Zhong SSW, Wu WC, Chu KC, Wong CY, Lau CY, Chen I, Chan NW, Zhang J. TISIDB: an integrated repository portal for tumor-immune system interactions. Bioinformatics. 2019;35:4200–2.
Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, Sun Y, Jacobsen A, Sinha R, Larsson E, Cerami E, Sander C, Schultz N. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signaling. 2013. https://doi.org/10.1126/scisignal.2004088.
Vasaikar SV, Straub P, Wang J, Zhang B. LinkedOmics: analyzing multi-omics data within and across 32 cancer types. Nucleic Acids Res. 2018;46:D956-d963.
Sun Q, Barz M, De Geest BG, Diken M, Hennink WE, Kiessling F, Lammers T, Shi Y. Nanomedicine and macroscale materials in immuno-oncology. Chem Soc Rev. 2019;48:351–81.
Liu XP, Ju L, Chen C, Liu T, Li S, Wang X. DNA methylation-based panel predicts survival of patients with clear cell renal cell carcinoma and its correlations with genomic metrics and tumor immune cell infiltration. Front Cell Dev Biol. 2020;8:572628.
Wei JH, Haddad A, Wu KJ, Zhao HW, Kapur P, Zhang ZL, Zhao LY, Chen ZH, Zhou YY, Zhou JC, Wang B, Yu YH, Cai MY, Xie D, Liao B, Li CX, Li PX, Wang ZR, Zhou FJ, Shi L, Liu QZ, Gao ZL, He DL, Chen W, Hsieh JT, Li QZ, Margulis V, Luo JH. A CpG-methylation-based assay to predict survival in clear cell renal cell carcinoma. Nat Commun. 2015;6:8699.
Wang J, Zhang Q, Zhu Q, Liu C, Nan X, Wang F, Fang L, Liu J, Xie C, Fu S, Song B. Identification of methylation-driven genes related to prognosis in clear-cell renal cell carcinoma. J Cell Physiol. 2020;235:1296–308.
Huang J, Chen W, Jie Z, Jiang M. Comprehensive analysis of immune implications and prognostic value of SPI1 in gastric cancer. Front Oncol. 2022;12:820568.
Wang YC, Wu YS, Hung CY, Wang SA, Young MJ, Hsu TI, Hung JJ. USP24 induces IL-6 in tumor-associated microenvironment by stabilizing p300 and β-TrCP and promotes cancer malignancy. Nat Commun. 2018;9:3996.
Gordon-Alonso M, Hirsch T, Wildmann C, van der Bruggen P. Galectin-3 captures interferon-gamma in the tumor matrix reducing chemokine gradient production and T-cell tumor infiltration. Nat Commun. 2017;8:793.
He Y, Jiang Z, Chen C, Wang X. Classification of triple-negative breast cancers based on Immunogenomic profiling. J Exp Clin Cancer Res. 2018;37:327.
Li Y, Fang M, Zhang J, Wang J, Song Y, Shi J, Li W, Wu G, Ren J, Wang Z, Zou W, Wang L. Hydrogel dual delivered celecoxib and anti-PD-1 synergistically improve antitumor immunity. Oncoimmunology. 2016;5:e1074374.
Chiu DK, Tse AP, Xu IM, Di Cui J, Lai RK, Li LL, Koh HY, Tsang FH, Wei LL, Wong CM, Ng IO, Wong CC. Hypoxia inducible factor HIF-1 promotes myeloid-derived suppressor cells accumulation through ENTPD2/CD39L1 in hepatocellular carcinoma. Nat Commun. 2017;8:517.
Kitamura T, Pollard JW. Therapeutic potential of chemokine signal inhibition for metastatic breast cancer. Pharmacol Res. 2015;100:266–70.
Zhu Y, Knolhoff BL, Meyer MA, Nywening TM, West BL, Luo J, Wang-Gillam A, Goedegebuure SP, Linehan DC, DeNardo DG. CSF1/CSF1R blockade reprograms tumor-infiltrating macrophages and improves response to T-cell checkpoint immunotherapy in pancreatic cancer models. Can Res. 2014;74:5057–69.
Wherry EJ. T cell exhaustion. Nat Immunol. 2011;12:492–9.
Wherry EJ, Kurachi M. Molecular and cellular insights into T cell exhaustion. Nat Rev Immunol. 2015;15:486–99.
Desbaillets I, Diserens AC, Tribolet N, Hamou MF, Van Meir EG. Upregulation of interleukin 8 by oxygen-deprived cells in glioblastoma suggests a role in leukocyte activation, chemotaxis, and angiogenesis. J Exp Med. 1997;186:1201–12.
Van Meir EG. Cytokines and tumors of the central nervous system. Glia. 1995;15:264–88.
Acknowledgements
None.
Funding
None.
Author information
Authors and Affiliations
Contributions
XZ and HY-F designed the study. HY-F, TW, DL, and ZL performed the experiment and data analysis. JL-Y, XH and YY performed clinical data analysis. HY-F and YH contributed to manuscript writing, reviewing, and revision. YH and XZ supervised the study. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
The protocol was approved by the ethics committee of the Chinese People’s Liberation Army (PLA) General Hospital in accordance with the Declaration of Helsinki.
Competing interests
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Additional file 1: Table S1.
The significant prognostic values of CpG in SPI1.
Additional file 2
: Table S2. Genes associated with SPI1 expression in ccRCC
Additional file 3
: Table S3. Clinical and immune phenotype data for the CheckMate cohorts.
Additional file 4: Fig. S1.
DNA methylation levels of SPI1 in ccRCC. (a). Methylation of SPI1 were lower in ccRCC bulk tissues than that in normal bulk tissues in the UALCAN Database. (b). Correlation analysis of SPI1 mRNA expression with SPI1 promoter methylation status using the cBioPortal database. (c-e).Promoter methylation level of SPI1 in ccRCC tumor bulk tissues of different tumor gade (c), tumor stage (d), and nodal metastasis status(e) using the UALCAN database. ccRCC, clear cell renal cell carcinoma; *, p < 0.05; **, p < 0.01; ***, p < 0.001; ns, non-significant. Fig. S2. Immune infiltrates in ccRCC are associated with the methylation status of SPI1. (a) Relationships between the methylation status of SPI1 and activated CD4 T cells, activated CD8 T cells, and activated dendritic cells in patients with ccRCC using the TISIDB database. (b-d) Relationships between the methylation status of SPI1 and immunostimulators (b), immunoinhibitors (c), and chemokines/receptors (d) in patients with using the TISIDB database. KIRC, kidney renal clear cell carcinoma; ACT_CD4, activated CD4+ T cells; ACT_CD8, activated CD8+ T cells; ACT_DC, activated dendritic cells. Fig. S3. Relationship between SPI1 expression and clinical benefit of immunotherapy in prospective clinical trials in ccRCC. (a) Patients with lower SPI1 expression benefit more from immunotherapy. CB, clinical benefit; NCB, no clinical benefit; ICB, intermediate clinical benefit.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Feng, H., Wang, T., Ye, J. et al. SPI1 is a prognostic biomarker of immune infiltration and immunotherapy efficacy in clear cell renal cell carcinoma. Discov Onc 13, 134 (2022). https://doi.org/10.1007/s12672-022-00592-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s12672-022-00592-0