Abstract
Primary brain cells cultured on flat surfaces, i.e., in a two-dimensional fashion, have a long history of use as an experimental model system in neuroscience research. However, it is questionable to which extent these cultured brain cells resemble their in vivo counterparts. Mainly, it has been claimed that the non-oxidative glucose metabolism reflected by lactate production is unphysiologically high. Furthermore, it is known that culturing in 2D alters the phenotype of cells. Here we present diphenylalanine peptide nanowires (PNWs) as a culturing substrate for primary neocortical neurons from mice. The topology of the PNWs leads to neuronal cultures developing in 2.5D environment and hence improved culturing conditions. We investigate the effect of different concentrations of PNWs and different cell densities of neurons on the culturing conditions. The neocortical neurons were examined through scanning electron microscopy in order to study the effect of PNW concentrations and neuron densities on the structural appearance of the cells. Then employing the optimal combination of neuron density and PNW concentration, the neurons were evaluated functionally and metabolically by comparison with neocortical neurons standard culturing methods in 2D. Specifically, we tested neuronal viability, capacity for vesicular release of neurotransmitter GABA, as well as oxidative and non-oxidative glucose metabolism. It was evident that neurons cultured on PNWs exhibited increased viability combined with an increased capacity for neurotransmitter release and a lower fraction of non-oxidative metabolism than neurons cultured in 2D. Hence, neocortical neurons cultured in 2.5D on PNWs appear to be healthier and less glycolytic than neurons cultured in 2D.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
Primary neuronal cell cultures have long been one of the key elements in neuroscience research. A wide range of neuronal cells have been utilized in order to obtain knowledge of the brain and its circuitry. Depending on the cell type, a variety of culturing protocols have been developed. Traditionally, neuronal cells are cultured in 2D on either polystyrene or glass. Most often, the cells are cultured using either culturing flasks or Petri dishes [1,2,3]. However, this is changing, and over the last decades, there has been an increased focus on developing systems that mimic the in vivo environment of cells. This is being achieved both by introducing lab-on-a-chip systems for culturing and by developing new culturing substrates. A common approach for creating more in vivo-like conditions is to provide the cells with a 3D environment, as it is known that culturing in 2D is more prone to alter the phenotype of cells. A simple and widely used method is to culture cells in a gel matrix [4,5,6]. However, having the cells in a matrix complicates post processing and analysis. By culturing in 2.5D on substrates with an alternative topology like for example membranes [7,8,9,10], nanostructures [11, 12], or even pillars [13,14,15,16], some of the benefits of 3D culturing can be obtained while providing the possibility of conventional post-culture handling. Both ordered and random structures have been demonstrated to affect cell growth. Unfortunately, these structures have not affected cell growth in a predictable manner as the various topologies influence the development of cells differently. This was observed not only in primary cultures of neurons which are known to be relatively fragile but also in more robust cell lines [13, 15, 17,18,19].
In the search for new culturing substrates, peptides are often encountered as a means to ensure temporally stable adhesion (important for implants) or enhance physical stability based on self-assembly properties. In relation to cell cultures, peptides are mainly used as a component in hydrogels or as a substrate modification [4,5,6, 20,21,22,23,24,25] but peptides also show potential within, e.g., drug delivery, biosensors [26,27,28,29], cleanroom fabrication [30], and for electrode functionalization [31, 32].
The peptide diphenylalanine has been shown to self-assemble into a wide range of structures, including but not limited to fibers and nanowires [26, 33]. Previously, cells such as chondrocytes and fibroblasts have been cultured on substrates based on modified diphenylalanine [34,35,36,37,38]. Unmodified diphenylalanine peptide nanowires (PNWs) have previously been shown to support and possibly improve neuronal cell line cultures [31, 39]. The PNWs provide an ordered surface at high concentrations (PNW grass) and a randomly ordered surface at lower concentrations [31]. In this paper, we visually examined the phenotype of primary cultures of neocortical neurons grown on various concentrations of diphenylalanine PNWs. Based on this, we select the most favorable concentration of PNWs and evaluate the functional and metabolic features of primary neocortical neurons cultured on a surface of unmodified PNWs. In parallel, neurons plated on poly-D-lysine-coated polystyrene in 2D served as control. As various studies show that cells thrive better on non-flat substrates [7, 19, 40,41,42,43,44], we hypothesize that neocortical neurons cultured on PNWs are functionally and metabolically superior to neurons cultured the traditional way in 2D.
2 Materials and Methods
2.1 Ethics Statement
All animal procedures were conducted in accordance with EU and Danish laws regulating animal experiments.
2.2 Materials
Chemicals for synthesis of peptide nanowires are diphenylalanine (BACHEM, Bubendorf, Switzerland), 1,1,1,3,3,3-hexafluoro-2-propanol (HFP; Fluka, Buchs, Switzerland), aniline (Sigma-Aldrich, St. Louis, MO, USA), and silica beads with orange indicator (Merck Millipore, Darmstadt, Germany). Pregnant mice of 15-day gestation were obtained from Harlan (Venray, the Netherlands). Fetal calf serum was purchased from Seralab Ltd. (Sussex, UK) while culture medium (Dulbecco’s minimal essential medium), poly-D-lysine (molecular weight > 300.000 g/mol), trypsin, DNase, cytosine arabinoside, N,N-dimethylformamide (DMF), and N-methyl-N-(tert-butyldimethylsilyl) trifluoroacetamide (MTBSTFA) were from Sigma-Aldrich (St. Louis, MO, USA). [U-13C] glucose was obtained from Isotec, Sigma-Aldrich (St. Louis, MO, USA); γ-vinyl-GABA (GVG, vigabatrin) was purchased from Tocris (Bristol, UK) and [3H] GABA was from Amersham Bioscience (Buckinghamshire, UK).
2.3 Synthesis of Self-Assembled Diphenylalanine Peptide Nanowires
Diphenylalanine PNWs were synthesized on cover glass employing a slight modification of the procedure previously described [31, 39, 45,46,47]. The procedure is illustrated in Fig. 1a. Briefly, solutions of 10, 25, 50, 75, 100, and 125 mg/ml diphenylalanine in HFP were prepared and added to the glass slide in an amount barely sufficient to cover the slide. Subsequently, the substrates were dried under anhydrous conditions in a vacuum desiccator for 15 min. The substrates were then placed in a glass container without a lid (Fig. 1b) and a second glass container was partly filled with aniline. The two containers were placed inside a third glass container which was covered by a lid. The entire ensemble was placed in a large glass Petri dish, and the space between the ensemble and the dish was filled with silica gel to absorb excess aniline vapor that might escape under the lid (Fig. 1b). Everything was wrapped in aluminum foil and placed in an oven at 120 °C for at least 10 h. After this, the package was unwrapped and the lid immediately removed from the large container to prevent condensation of aniline. It should be noted that the high concentration of PNWs will result in vertically aligned wires (PNW grass) while low concentrations will result in wires that are scattered across the surface with a random orientation and not necessarily covering the entire surface [31].
Preparation of peptide nanowires (PNWs). a Illustration of PNW synthesis. b Setup for PNW synthesis. One container holds the glass substrates with a monolayer of dry diphenylalanine and a second holds aniline for creating aniline vapor. The containers are placed in a larger container with a lid to create a closed system.
2.4 Preparation of Primary Neocortical Neuron Cultures
Neocortical neurons were isolated and cultured as previously described [48, 49]. Briefly, pregnant mice of 15-day gestation were sacrificed by cervical dislocation, and the embryos were immediately dissected from the amnions under aseptic conditions. Cerebral cortices were isolated from each embryo and the tissue was diced using a razor blade. Following trypsinization (0.2 mg/ml trypsin, 37 °C, 10 min), the cells were triturated in a solution containing DNase (75 IU/ml) and trypsin inhibitor from soybean (0.53 mg/ml). The cells were suspended in a slightly modified Dulbecco’s minimal essential medium (DMEM) containing 24.5 mM KCl, 12 mM glucose, and 10% fetal calf serum. Subsequently, the cells were seeded in 35-mm Petri dishes with or without a cover glass insert containing PNWs. Prior to culturing, all Petri dishes (with and without PNWs) were coated with poly-D-lysine (50 mg/L) overnight at room temperature. The neocortical neuron cultures were kept at 37 °C in a humidified atmosphere and used for experiments after 7–8 days in vitro. On the second day of culturing, cytosine arabinoside was added (20 μM; final concentration) to prevent astrocyte proliferation. Further, on days 2 and 5 the culture medium was supplemented with glucose to a minimum concentration of 12 mM.
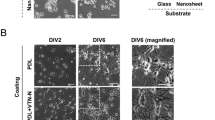
For the initial screening, two different densities of cells (1.75 × 106 cells per ml and 2.5 × 106 cells per ml) were cultured on 8 different substrates, i.e., two reference substrates without PNWs (polystyrene and glass) and 6 substrates (glass) with 10, 25, 50, 75, 100, and 125 mg/ml PNWs, respectively. The neuronal cells cultured on the different substrates were evaluated visually by qualitatively assessing three factors, i.e., homogeneity of cell coverage, cell connectivity, and morphological appearance (Fig. 2).
2.5 Fixation of Cell Cultures
In order to visually inspect the phenotype of the cells, the cell cultures were fixated using 2% glutaraldehyde. The culture medium was removed from the Petri dish and the neurons were washed twice with phosphate buffered saline (PBS). The cells were subsequently fixated in 2% glutaraldehyde dissolved in 0.1 M PBS for 1 h. The fixated neurons on cover glass were bathed 2 times 5 min in PBS and then 10 min in sterile water. The cells were dehydrated in a series of ethanol baths (50-60-70-80-90-100%, 5 min each) and then air dried.
2.6 Scanning Electron Microscopy
The fixated cells were inspected visually using a FEI Quanta 200 ESEM FEG scanning electron microscope. The samples were placed on aluminum pin stubs (Agar Scientific, Essex, United Kingdom) using Leit Adhesive Carbon Tabs (Agar Scientific, Essex, United Kingdom). The microscope was operated under low vacuum conditions using a large field detector. Water vapor was used to visualize the low/non-conductive samples. The samples were inspected from the top using an angle perpendicular to the cell layer.
2.7 Viability Test
After 7 days in vitro, neuronal viability was tested by assessing lactate dehydrogenase (LDH) activity in cell extracts and in the culture medium, i.e., intra- and extracellularly. This method is based on the fact that the cytosolic enzyme LDH is able to traverse the membrane only when the membrane is disrupted, i.e., in dead cells [50]. Moreover, we tested neuronal robustness by assessing neuronal viability following a challenging event, namely following incubation for 24 h in a fresh medium devoid of fetal calf serum. Again, LDH activity was determined both intra- and extracellularly. LDH activity in cell extracts and media was measured employing a colorimetric LDH assay kit (Abcam, Cambridge, UK; product number: ab102526), and the assay was conducted according to the manufacturer’s instructions. The viability was calculated as the intracellular LDH activity relative to total LDH activity (the sum of intra- and extracellular LDH activity).
2.8 Vesicular Release of Neurotransmitter GABA
Neocortical neurons cultured on PNWs or in 2D on polystyrene were pre-incubated in the presence of the GABA transaminase inhibitor γ-vinyl-GABA (GVG; Vigabatrin) for 30 min prior to loading of [3H] GABA for another 30 min. Pre-incubation with GVG was necessary to avoid metabolism of the radiolabeled GABA [51] as this would confound the subsequent data interpretation. After the loading period, the cell cultures were transferred to a superfusion paradigm as described by Drejer et al. 1987 [51] and superfused with HEPES buffer containing in mM: HEPES, 10; NaCl, 135; KCl, 5; CaCl2, 1; MgSO4, 0.6; and glucose, 2.5. The flow was 2 ml/min. After 10 min of equilibration in this medium, depolarization was induced by switching the medium to a HEPES buffer containing 55 mM K+ (with an equimolar reduction in Na+) for 30 s every 4 min for a total of 8 stimulations. The superfusion medium was continuously collected in fractions of 30 s, i.e., 1 ml/fraction. At the end of an experiment, the cells were extracted using 70% ethanol, and the cell extracts as well as all fractions were analyzed for radioactivity by scintillation counting [52]. Vesicular release of GABA was calculated by dividing the radioactivity in each fraction by the total amount of intracellular radioactivity at that time, i.e., the sum radioactivity of subsequent fractions plus the radioactivity in the cell extract at the end of the experiment. The release curves obtained in each batch of neuronal cultures [3, 4] were averaged, and the resulting curve was used to determine the area under the curve (AUC) for each peak. These values were subsequently normalized to the control peaks generated in the same batch of neuronal cultures.
2.9 Incubation Experiments to Investigate Glucose Metabolism
Culture medium from cells grown on PNWs or polystyrene was removed; the cells were washed once with 37 °C PBS and incubated in a modified DMEM without serum containing 2.5 mM [U-13C] glucose for 30 min. Subsequently, the incubation medium was collected, and the cell layer was washed once with ice cold PBS. The cells were extracted employing 2 × 500 μl ice cold 70% ethanol and scraped off the substrate using a rubber policeman. When PNWs were used as substrate, these were scraped off simultaneously. Following centrifugation at 20,000g for 10 min at 4 °C the supernatant was transferred to an Eppendorf tube and lyophilized before HPLC and GC-MS analyses while the pellet was used for protein determination.
2.10 Protein Determination
The pellets consisting of cell remnants and in some cases PNWs were left at room temperature overnight to dissolve in 1 M KOH. While the cell remnants were soluble in KOH, the PNWs were not. Following a short spin at 600 rpm for 30 s at room temperature, the supernatant was removed and kept at − 20 °C until analysis for protein determination. The protein content was measured employing a Bicinchoninic acid (BCA) assay (Thermo Scientific, Rockford, IL) using bovine serum albumin (BSA) as standard and absorbance was determined at 550 nm.
2.11 HPLC Analysis
The lyophilized cell extracts were resuspended in water, and an aliquot was used to determine the content of alanine, glutamate, GABA, and aspartate intracellularly. The amino acids were pre-column derivatized with the fluorogenic orthophthalaldehyde (OPA) in 0.4 M borate buffer (Agilent 5061-3339; pH 10.2) and separated by reversed phase HPLC using a C18 column (Zorbax Eclipse Plus 4.6150 mm, particle size 3.5 μm; 959963-902, Agilent Technologies, Santa Clara, CA, USA). A mobile phase gradient was used employing mobile phase A (10 mM NaH2PO4, 10 mM Na2B4O7, 0.5 mM NaN3; pH 8.2) and mobile phase B (acetonitrile:methanol:milli-Q water; 45:45:10 (v:v:v)). The percentage of mobile phase B increased linearly from 2 to 57% in 30 min, and from 57 to 100% in 0.1 min. At time 33.6 min, the percent of mobile phase B returned to 2% and was maintained at that level. The total run time was 35 min and the flow of the mixed mobile phases was kept at 1.5 ml/min. A standard curve containing the relevant amino acids in concentrations from 5 to 300 μM was used to determine amino acid content in the sample and this was related to the protein content.
2.12 GC-MS Analysis
Following lyophilization of cell extracts and reconstitution in water, the incorporation of 13C from glucose into relevant metabolites (lactate, alanine, glutamate, GABA, and aspartate) was determined employing GC-MS as previously described by Walls et al. 2014 [49]. Briefly, an aliquot of the reconstituted cell extracts was acidified using HCl (pH 1-2) and dried under a nitrogen flow. The metabolites were extracted into an organic phase using successive steps of ethanol and benzene addition. Following evaporation to complete dryness under nitrogen, the metabolites were derivatized with MTBSTFA containing 1% v/v tertbutyldimethylchlorosilane in the presence of N,N-dimethylformamide (DMF, Mawhinney et al. 1986) [53]. Analyses were performed on an Agilent Technologies 7820A chromatograph, J&W GC column HP-5MS, parts no. 19091S-433 coupled to a mass spectrometer (Agilent Technologies 5977E). All data were corrected for natural abundance determined from an unlabeled standard containing relevant metabolites and which was analyzed in the same GC-MS sequence. The enrichment of 13C in metabolites was calculated as described by Walls et al. 2014 and Biemann 1962 [49, 54].
2.13 Metabolism of [U-13C]Glucose
Each molecule of [U-13C] glucose can, via glycolysis, be metabolized to two molecules of [U-13C] pyruvate. Due to the high activity of the enzymes lactate dehydrogenase and alanine aminotransferase, the pyruvate pool is in equilibrium with the pools of lactate and alanine. As the pool of pyruvate is small and below the detection limit of the GC-MS instrument, the 13C labeling in lactate and alanine reflects the labeling in pyruvate. [U-13C] pyruvate may alternatively be converted to [1,2-13C] acetyl co-enzyme A which is introduced into the TCA cycle by condensation with oxaloacetate to form double-labeled (M + 2) citrate. This is further converted to double-labeled α-ketoglutarate which may give rise to double-labeled (M + 2) glutamate. The 13C labeling in glutamate reflects that in α-ketoglutarate due to the high activity of aminotransferases exceeding that of the TCA cycle by several fold [55]. Glutamate is precursor for GABA synthesis and hence double-labeled (M + 2) glutamate can give rise to double-labeled (M + 2) GABA. Alternatively, double-labeled (M + 2) α-ketoglutarate may be metabolized in the TCA cycle to form double-labeled (M + 2) oxaloacetate which may be transaminated to double-labeled (M + 2) aspartate by aspartate aminotransferase.
2.14 Data Analysis
The culturing conditions for neurons on PNWs were qualitatively determined from the initial scanning electron microscope inspections, which indicated that a density of 1.75 × 106 cells per ml cultured on 50 mg/ml PNWs was optimal. Statistically significant differences between data obtained from neurons cultured on PNWs under these conditions and those from controls cultured in 2D on polystyrene were determined employing an unpaired Student’s t test. Graph Pad Prism 6 was used to identify significant differences and data were taken to be statistically different when P < 0.05. Data were excluded from the data set when identified as significant outliers employing Rout’s outlier test. All results are presented as averages ± standard error of the mean (SEM).
3 Results and Discussion
Neurons plated at densities of 1.75 × 106 cells per ml and 2.5 × 106 cells per ml were morphologically evaluated by scanning electron microscopy. For each cell culture, 5 to 10 spots were visually inspected following fixation with glutaraldehyde. The density and appearance of cells varied across the culture dish, and the results presented in Fig. 2 show the general trend observed. At a density of 1.75 × 106 cells per ml, the neurons appeared morphologically healthier, and the cell distribution on the surfaces was more homogeneous compared to cells plated at a density of 2.5 × 106 cells per ml. In addition, plating with this higher density resulted in the cells being washed off during the fixation procedure, and hence the lower cell density was chosen for the subsequent characterization of neurons cultured on PNWs and polystyrene, respectively.
From the visual inspection, we observed that in general, neurons cultured on peptide-covered surfaces appeared healthier than those cultured in 2D independent of the surface material (glass or plastic). The neurons cultured on bare glass cover slips were often migrated into clusters (Fig. 2b) whereas the cells on both plastic and peptide-covered glass exhibited a more homogeneous distribution (Fig. 2a). Neocortical neurons cultured on low concentrations of PNWs (10 mg/ml; Fig. 2d) resemble those cultured in 2D on polystyrene (Fig. 2b). However, once the layer of PNWs becomes confluent (50 mg/ml and 100 mg/ml), the health status of the neurons seems to improve as observed by increased homogeneity and connectivity (Fig. 2a, c).
It was noted that many neurons did not retain their shape throughout the fixation procedure, resulting in a smeared appearance (Fig. 2f as opposed to Fig. 2e, where the cell soma is retained). This is a common undesired side effect of the fixation-dehydration procedure and was observed, irrespective of cell density and peptide concentration. Compared to previous work conducted by other research groups, our neuron cultures appear healthy with a well-developed neurite network [40,41,42,43,44]. However, as the dimensions of the PNWs are of the same order of magnitude as the neurites, it is difficult to distinguish the full extent of the network (Fig. 2c, e).
Altogether, conclusion based on visual inspection using scanning electron microscopy is that primary neocortical neurons seem to thrive better on a surface covered by PNWs compared to that of a flat 2D glass or polystyrene surface. A density of 1.75 × 106 cells per ml cultured on 50 mg/ml PNWs appeared (at least qualitatively) to provide the most favorable culturing conditions, and hence this combination was used for further characterization of neocortical neurons cultured on PNWs.
The neocortical neurons grown using this selected procedure was then further investigated for improved function, with regard to viability, functionality, and metabolic activity. Sister neurons cultured the traditional way in 2D on polystyrene served as control.
Viability was assessed by measuring LDH activity intra- and extracellularly and is reported as the percent of intracellular LDH activity relative to total LDH activity (the sum of intra- and extracellular LDH activity). It was observed that in the control condition, approximately 95% of the neocortical neurons cultured on PNWs were viable following 1 week in vitro compared to approximately 86% of those cultured in 2D on polystyrene (Fig. 3; control).
Cell viability, Cell viability was tested in neurons cultured on polystyrene (black) and on peptide nanowires (PNWs; white) by measuring LDH activity intra- and extracellularly following 7 days in vitro (control) or after 7 days in vitro and subsequently incubation for 24 h in a fresh medium devoid of FCS (-FCS). Data presented are average ± SEM, n = 4–6 from two different batches of neuronal cultures. Differences between neurons cultured on polystyrene and on PNWs were determined by a Student’s t test and an asterisk (*) indicates statistical significance (P < 0.05).
This slightly higher viability for neurons cultured on PNWs corresponding to an increase of only about 10% may not seem noteworthy although statistically significant (P = 0.0138). However, the mortality ratio calculated as the percent extracellular to total LDH activity is reduced from ~ 14 to ~ 5%, i.e., a remarkable reduction in mortality of about 60%, when neurons are grown on PNWs. This greater viability when PNWs are used as substrate for culturing is compatible with the visual inspections from the scanning electron microscope proposing that neurons cultured on PNWs thrive better than those cultured in 2D. Although not significant, the same tendency is observed when the cells were challenged by changing the medium to a fresh medium devoid of fetal calf serum (Fig. 3; -FCS) for 24 h.
It should be noted that the control condition reflects cell death during the 7-day culturing period whereas the -FCS condition reflects cell death over a 24-h period following a medium change. Accordingly, the percent viability calculated for the two conditions cannot be directly compared, and the similar percentages observed in the control and the -FCS conditions do not reflect equal intracellular LDH activity. Actually, the intracellular LDH activity in the -FCS condition is only about one-third of that observed following 7 days of culturing (results not shown), confirming the harmful effects of removing fetal calf serum.
Functionally, the neurons cultured on PNWs were tested with regard to their capacity to release preloaded [3H] GABA upon stimulation induced by an elevated K+ concentration. The release curves for neurons cultured on PNWs and on polystyrene, respectively, were calculated as the fractional radioactivity in each superfusion fraction relative to the intracellular radioactivity at the given time (Fig. 4a). To visualize any differences in AUC between neurons cultured on PNWs and those on polystyrene, the baselines were in both cases adjusted to the same average, in this case, 0.05. The release curves were analyzed for AUC for each peak reflecting vesicular release of neurotransmitter GABA, i.e., neuronal function, and normalized letting the control represent 100%. The vesicular GABA release from neurons cultured on PNWs was significantly higher compared to that observed in controls (Fig. 4b). Again, this supports the hypothesis that neurons appear to thrive better on PNWs compared to when cultured in 2D. Moreover, it is generally accepted that the cytosolic and maybe more importantly, the nerve ending concentration of GABA determines the efficacy of inhibition [56,57,58]. Although the total amount of GABA in the neurons cultured on PNWs is similar to that of neurons cultured on polystyrene (Fig. 5a), it is likely that GABA is differentially distributed among the vesicular and the metabolic pool of GABA in the two settings. It has previously been demonstrated that the vesicular and the metabolic pools of GABA are associated with different mitochondria [59], and it may be hypothesized that a higher fraction of mitochondria is associated with the vesicular pool in the neurons grown on PNWs.
Release of neurotransmitter GABA. a A representative profile of [3H] GABA release from neocortical neurons cultured on peptide nanowires (PNWs; gray) or on polystyrene (CTRL; black) following repetitive depolarization. Each release curve represents the average from 3 to 4 culture dishes from the same batch of neuronal cultures. Baselines are adjusted to 0.05 in order to be able to compare AUCs visually. b From the release curves, the AUC was determined for each peak from neurons cultured on polystyrene (CTRL; black) and PNWs (white) and normalized letting the control situation represent 100%. Data presented are average ± SEM, n = 15–16 peaks from two different batches of neuronal cultures. Differences between neurons cultured on polystyrene and on PNWs were determined by a Student’s t test and an asterisk (*) indicates statistical significance (P < 0.05).
Glucose metabolism. Neocortical neurons cultured on polystyrene (black) or peptide nanowires (PNWs; white) were incubated in medium containing 2.5 mM [U-13C] glucose for 30 min. Subsequently, cell extracts were analyzed for a the amounts of amino acids, b the percentage triple (M + 3)-labeled lactate and alanine, and c the percentage double (M + 2) labeling in glutamate, GABA, and aspartate, and d) the metabolic pathways glycolysis and TCA cycle metabolism and the labeling patters in metabolites labeled from [U-13C] glucose. Data presented are average ± SEM, n = 7–9 from two different batches of neuronal cultures. Differences between neurons cultured on PS and on PNW were determined by a Student’s T test and an asterisk (*) indicates statistical significance (P < 0.05). Ala, alanine; Lac, lactate; Glu, glutamate; Asp, aspartate
Finally, the neurons cultured on PNWs were characterized with regard to the main metabolic pathways used for metabolism of glucose, i.e., glycolysis and TCA cycle metabolism. The neuronal cultures were incubated in medium containing 6 mM [U-13C] glucose for 30 min. Following an experiment, the ethanol cell extracts were analyzed for the content of alanine, glutamate, aspartate, and GABA using HPLC and for the percent 13C labeling in lactate, alanine, glutamate, aspartate, and GABA employing GC-MS. While the GC-MS data reveals the 13C labeling incorporated into metabolites during the 30-min incubation period, the HPLC data reports metabolite pool sizes generated over a longer time period.
Glycolytic activity was assessed by the percent uniformly (M + 3) labeled lactate and alanine generated from metabolism of [U-13C] glucose (Fig. 5b). The percent of uniformly labeled (M + 3) lactate and alanine was markedly reduced by about 90% and 75%, respectively, in neurons cultured on PNWs compared to those grown on polystyrene (Fig. 5b). This clearly demonstrates that non-oxidative glucose metabolism was reduced in neurons cultured on PNWs.
The extent of [U-13C] glucose being metabolized via glycolysis and followed by metabolism in the TCA cycle was determined by the extent of double labeling (M + 2) in glutamate, GABA, and aspartate. It was evident that the percentage of these compounds being double labeled (M + 2) from [U-13C] glucose was unaltered in neurons cultured on PNW compared to those grown on polystyrene (Fig. 5C), suggesting an unaltered oxidative metabolism via the TCA cycle. In combination with the attenuated glycolytic activity, this implies that overall glucose metabolism is lower in neurons cultured on PNWs than in neurons grown on polystyrene.
Altogether, these data propose that oxidative metabolism of glucose via the TCA cycle accounts for a larger fraction of total glucose metabolism in neurons cultured on PNWs than in control cultures. This may reflect a higher efficiency of the mitochondria in neurons cultured on PNWs, i.e., re-oxidation of NADH and FADH2 in the electron transport chain results in a greater ATP production. Overall, this improves cellular bioenergetics by enhancing the amount of ATP generated per molecule of glucose being oxidized. The ATP-synthetase (Complex V) is driven by a proton gradient across the inner mitochondrial membrane generated in the electron transferring entities complex I, III, and IV. It has been proposed that these three complexes are able to reversibly associate into a supercomplex that organizes electron flux to optimize the use of available substrates [60]. Hence, it may be hypothesized that the formation of supercomplexes is more frequent in neurons cultured on PNWs leading to an improved mitochondrial efficiency.
Alternatively, it may be speculated that a greater mitochondrial efficiency in the neurons cultured on PNWs arises as a consequence of reduced mitochondrial uncoupling. Mitochondrial uncoupling describes the situation when the proton gradient across the inner mitochondrial membrane is degraded circumventing complex V and ATP production. The consequence is an increase in glucose consumption in order to maintain ATP production. Likely, mitochondrial uncoupling also leads to an increase in non-oxidative glucose metabolism, i.e., lactate production. The lactate dehydrogenase reaction serves to re-oxidize NADH thereby ensuring continuance of glycolytic flux and ATP production independent of mitochondrial function. Once again, our data point to the fact that neurons cultured on PNWs thrive better than neurons cultured on polystyrene in 2D owing to more efficient mitochondria. This is compatible with the finding that mitochondrial dysfunction comprises a key element in neurodegenerative diseases characterized by progressive loss of neuronal function and structure [61,62,63].
In addition, the amount of alanine in neurons cultured on PNWs was significantly higher compared to that in neurons cultured on polystyrene. However, the increase in alanine content was not associated with a similar increase in the percent of uniformly labeled alanine (M + 3), which was in fact substantially reduced in neurons cultured on PNWs (Fig. 5b). This suggests a slower turnover of the alanine pool in neurons cultured on PNWs. Degradation of alanine can support continuance of TCA cycle activity by supplying pyruvate which can enter the TCA cycle following conversion to acetyl co-enzyme A. This may be of particular importance when a large fraction of the pyruvate generated from glucose is utilized for lactate production, i.e., during non-oxidative glucose metabolism, as observed in neurons cultured on polystyrene. Again, the data support the hypothesis that neurons cultured on PNWs exhibit a higher bioenergetic efficiency than neurons cultured on polystyrene in 2D.
4 Conclusions
We have demonstrated that neocortical neurons experience better growth condition cultured on PNWs; we found an optimal density of 1.75 × 106 cells per ml cultured on 50 mg/ml PNWs provided the most favorable culturing conditions for neocortical neurons. The neocortical neurons cultured on PNWs appear healthier and exhibit enhanced viability, a higher neurotransmitter release, and a lower fraction of non-oxidative glucose metabolism. The fact that the neocortical neurons cultured on PNWs display a higher fraction of oxidative metabolism is likely to reflect improvements in mitochondrial bioenergetics compared to neurons cultured in 2D on polystyrene. In addition, an increased neurotransmitter release, which is a key indicator of neuronal function in combination with increased viability, implies that neurons cultured in 2.5D on PNWs provide a neuronal model system that provides healthier, neurons than those cultured traditionally in 2D on polystyrene.
References
Ahlemeyer, B., & Baumgart-Vogt, E. (2005). Optimized protocols for the simultaneous preparation of primary neuronal cultures of the neocortex, hippocampus and cerebellum from individual newborn (P0.5) C57Bl/6J mice. Journal of Neuroscience Methods, 149(2), 110–120.
Du, F., Qian, Z. M., Zhu, L., Wu, X. M., Qian, C., Chan, R., & Ke, Y. (2010). Purity, cell viability, expression of GFAP and bystin in astrocytes cultured by different procedures. Journal of Cellular Biochemistry, 109(1), 30–37.
Pacifici, M., & Peruzzi, F. (2012). Isolation and culture of rat embryonic neural cells: A quick protocol. JoVE, 63, e3965.
Holmes, T. C., de Lacalle, S., Su, X., Liu, G., Rich, A., & Zhang, S. (2000). Extensive neurite outgrowth and active synapse formation on self-assembling peptide scaffolds. Proceedings of the National Academy of Sciences, 97(12), 6728–6733.
Leipzig, N. D., Wylie, R. G., Kim, H., & Shoichet, M. S. (2011). Differentiation of neural stem cells in three-dimensional growth factor-immobilized chitosan hydrogel scaffolds. Biomaterials, 32(1), 57–64.
Chiu, Y.-W., Chen, W.-P., Su, C.-C., Lee, Y.-C., Hsieh, P.-H., & Ho, Y.-L. (2014). The arrhythmogenic effect of self-assembling nanopeptide hydrogel scaffolds on neonatal mouse cardiomyocytes. Nanomedicine: Nanotechnology, Biology and Medicine, 10(5), e1065–e1073.
Bruggemann, D. (2013). Nanoporous Aluminium Oxide Membranes as Cell Interfaces. Journal of Nanomaterials, 2013, 18.
Alatraktchi, F., Bakmand, T., Dimaki, M., & Svendsen, W. (2014). Novel membrane-based electrochemical sensor for real-time bio-applications. Sensors, 14(11), 22128.
Bakmand, T.; Troels-Smith, A. R.; Dimaki, M.; Nissen, J. D.; Andersen, K. B.; Sasso, L.; Waagepetersen, H. S.; Gramsbergen, J. B.; Svendsen, W. E., Fluidic system for long-term in vitro culturing and monitoring of organotypic brain slices. Biomedical Microdevices 2015, 17 (4).
Svendsen, W. E.; Al Atraktchi, F. A. Z.; Bakmand, T.; Waagepetersen, H.; Dimaki, M.; Ieee, Novel culturing platform for brain slices and neuronal cells. In 2015 37th Annual International Conference of the Ieee Engineering in Medicine and Biology Society, 2015; pp 346–349.
Svendsen, W. E., Castillo-Leon, J., Lange, J. M., Sasso, L., Olsen, M. H., Abaddi, M., Andresen, L., Levinsen, S., Shah, P., Vedarethinam, I., & Dimaki, M. (2011). Micro and nano-platforms for biological cell analysis. Sensors and Actuators a-Physical, 172(1), 54–60.
Belu, A., Yilmaz, M., Neumann, E., Offenhaeusser, A., Demirel, G., & Mayer, D. (2018). Asymmetric, nano-textured surfaces influence neuron viability and polarity. Journal of Biomedical Materials Research Part A, 106(6), 1634–1645.
Dowell-Mesfin, N. M., Abdul-Karim, M. A., Turner, A. M. P., Schanz, S., Craighead, H. G., Roysam, B., Turner, J. N., & Shain, W. (2004). Topographically modified surfaces affect orientation and growth of hippocampal neurons. Journal of Neural Engineering, 1(2), 78.
Tania, L., Fabrizia, C., Francesco, G., Roberto, M., Roberta, R., Andrea, B., Marco, D. M., Maria, P. E., Stefania, S., Fabio, B., & Enzo, D. F. (2013). Nanostructured superhydrophobic substrates trigger the development of 3D neuronal networks. Small, 9(3), 402–412.
Ankam, S., Suryana, M., Chan, L. Y., Moe, A. A. K., Teo, B. K. K., Law, J. B. K., Sheetz, M. P., Low, H. Y., & Yim, E. K. F. (2013). Substrate topography and size determine the fate of human embryonic stem cells to neuronal or glial lineage. Acta Biomaterialia, 9(1), 4535–4545.
Toma, M.; Belu, A.; Mayer, D.; Offenhaeusser, A., Flexible gold nanocone array surfaces as a tool for regulating neuronal behavior. Small 2017, 13 (24).
Craighead, H. G., James, C. D., & Turner, A. M. P. (2001). Chemical and topographical patterning for directed cell attachment. Current Opinion in Solid State and Materials Science, 5(2), 177–184.
Micholt, L., Gärtner, A., Prodanov, D., Braeken, D., Dotti, C. G., & Bartic, C. (2013). Substrate topography determines neuronal polarization and growth in vitro. PLoS One, 8(6), e66170.
Wen-Ta, S., Yung-Feng, L., Tai-Wei, W., Bo-Jeng, W., & Yu-Yin, S. (2013). Microgrooved patterns enhanced PC12 cell growth, orientation, neurite elongation, and neuritogenesis. Journal of Biomedical Materials Research Part A, 101A(1), 185–194.
Massia, S. P., & Hubbell, J. A. (1990). Covalently attached Grgd on polymer surfaces promotes biospecific adhesion of mammalian-cells. Annals of the New York Academy of Sciences, 589, 261–270.
Massia, S. P., & Hubbell, J. A. (1991). Human endothelial-cell interactions with surface-coupled adhesion peptides on a nonadhesive glass substrate and 2 polymeric biomaterials. Journal of Biomedical Materials Research, 25(2), 223–242.
Mann, B. K., & West, J. L. (2002). Cell adhesion peptides alter smooth muscle cell adhesion, proliferation, migration, and matrix protein synthesis on modified surfaces and in polymer scaffolds. Journal of Biomedical Materials Research, 60(1), 86–93.
Jeschke, B., Meyer, J., Jonczyk, A., Kessler, H., Adamietz, P., Meenen, N. M., Kantlehner, M., Goepfert, C., & Nies, B. (2002). RGD-peptides for tissue engineering of articular cartilage. Biomaterials, 23(16), 3455–3463.
Schackel, T., Kumar, P., Gunther, M., Liu, S. W., Brunner, M., Sandner, B., Puttagunta, R., Muller, R., Weidner, N., & Blesch, A. (2019). Peptides and astroglia improve the regenerative capacity of alginate gels in the injured spinal cord. Tissue Engineering Part A, 25(7–8), 522–537.
Haagdorens, M., Cepla, V., Melsbach, E., Koivusalo, L., Skottman, H., Griffith, M., Valiokas, R., Zakaria, N., Pintelon, I., & Tassignon, M. J. (2019). In vitro cultivation of limbal epithelial stem cells on surface-modified crosslinked collagen scaffolds. Stem Cells International, 17.
Yan, X., Zhu, P., & Li, J. (2010). Self-assembly and application of diphenylalanine-based nanostructures. Chemical Society Reviews, 39(6), 1877–1890.
Suhito, I. R., Lee, W., Baek, S., Lee, D., Min, J., & Kim, T. H. (2019). Rapid and sensitive electrochemical detection of anticancer effects of curcumin on human glioblastoma cells. Sensors and Actuators B-Chemical, 288, 527–534.
Faya, M., Kalhapure, R. S., Dhumal, D., Agrawal, N., Omolo, C., Akamanchi, K. G., & Govender, T. (2019). Antimicrobial cell penetrating peptides with bacterial cell specificity: pharmacophore modelling, quantitative structure activity relationship and molecular dynamics simulation. Journal of Biomolecular Structure & Dynamics, 37(9), 2370–2380.
Tian, L., Tao, L., Li, H. Y., Zhao, S. L., Zhang, Y. F., Yang, S., Xue, J. F., & Zhang, X. F. (2019). Hollow mesoporous carbon modified with cRGD peptide nanoplatform for targeted drug delivery and chemo-photothermal therapy of prostatic carcinoma. Colloids and Surfaces a-Physicochemical and Engineering Aspects, 570, 386–395.
Andersen, K. B.; Castillo-Leon, J.; Bakmand, T.; Svendsen, W. E., Alignment and use of self-assembled peptide nanotubes as dry-etching mask. Jpn J Appl Phys 2012, 51 (6).
Sasso, L., Vedarethinam, I., Emnéus, J., Svendsen, W. E., & Castillo-Leóon, J. (2012). Self-assembled diphenylalanine nanowires for cellular studies and sensor applications. Journal of Nanoscience and Nanotechnology, 12(4), 3077–3083.
Castillo, J. J., Svendsen, W. E., Rozlosnik, N., Escobar, P., Martineza, F., & Castillo-Leon, J. (2013). Detection of cancer cells using a peptide nanotube-folic acid modified graphene electrode. Analyst, 138(4), 1026–1031.
Castillo-Leon, J., Rodriguez-Trujillo, R., Gauthier, S., Jensen, A. C. O., & Svendsen, W. E. (2011). Micro-“factory” for self-assembled peptide nanostructures. Microelectronic Engineering, 88(8), 1685–1688.
Liebmann, T., Rydholm, S., Akpe, V., & Brismar, H. (2007). Self-assembling Fmoc dipeptide hydrogel for in situ 3D cell culturing. BMC Biotechnology, 7, 88–88.
Jayawarna, V., Smith, A., Gough, J. E., & Ulijn, R. V. (2007). Three-dimensional cell culture of chondrocytes on modified di-phenylalanine scaffolds. Biochemical Society Transactions, 35(3), 535–537.
Jayawarna, V., Richardson, S. M., Hirst, A. R., Hodson, N. W., Saiani, A., Gough, J. E., & Ulijn, R. V. (2009). Introducing chemical functionality in Fmoc-peptide gels for cell culture. Acta Biomaterialia, 5(3), 934–943.
Zhou, M., Smith, A. M., Das, A. K., Hodson, N. W., Collins, R. F., Ulijn, R. V., & Gough, J. E. (2009). Self-assembled peptide-based hydrogels as scaffolds for anchorage-dependent cells. Biomaterials, 30(13), 2523–2530.
Zhou, M., Ulijn, R. V., & Gough, J. E. (2014). Extracellular matrix formation in self-assembled minimalistic bioactive hydrogels based on aromatic peptide amphiphiles. Journal of Tissue Engineering, 5, 2041731414531593.
Taskin, M. B., Sasso, L., Dimaki, M., Svendsen, W. E., & Castillo-León, J. (2013). Combined cell culture-biosensing platform using vertically aligned patterned peptide nanofibers for cellular studies. ACS Applied Materials & Interfaces, 5(8), 3323–3328.
Lovat, V., Pantarotto, D., Lagostena, L., Cacciari, B., Grandolfo, M., Righi, M., Spalluto, G., Prato, M., & Ballerini, L. (2005). Carbon nanotube substrates boost neuronal electrical signaling. Nano Letters, 5(6), 1107–1110.
Sann, H., Zdobnova, I., Kreutz, M. R., & Zuschratter, W. (2007). Correlative CLSM-SEM microscopy of synaptic proteins in primary neuronal cultures. Microscopy and Microanalysis, 13(S03), 194–195.
Mazzatenta, A., Giugliano, M., Campidelli, S., Gambazzi, L., Businaro, L., Markram, H., Prato, M., & Ballerini, L. (2007). Interfacing neurons with carbon nanotubes: electrical signal transfer and synaptic stimulation in cultured brain circuits. The Journal of Neuroscience, 27(26), 6931–6936.
Ojovan, S. M., McDonald, M., Rabieh, N., Shmuel, N., Erez, H., Nesladek, M., & Spira, M. E. (2014). Nanocrystalline diamond surfaces for adhesion and growth of primary neurons, conflicting results and rational explanation. Frontiers in Neuroengineering, 7, 17.
Cesca, F., Limongi, T., Accardo, A., Rocchi, A., Orlando, M., Shalabaeva, V., Di Fabrizio, E., & Benfenati, F. (2014). Fabrication of biocompatible free-standing nanopatterned films for primary neuronal cultures. RSC Advances, 4(86), 45696–45702.
Jungki, R., & Beum, P. C. (2008). High-temperature self-assembly of peptides into vertically well-aligned nanowires by aniline vapor. Advanced Materials, 20(19), 3754–3758.
Jungki, R., & Beum, P. C. (2009). Synthesis of diphenylalanine/polyaniline core/shell conducting nanowires by peptide self-assembly. Angewandte Chemie International Edition, 48(26), 4820–4823.
Jungki, R., & Beum, P. C. (2010). High stability of self-assembled peptide nanowires against thermal, chemical, and proteolytic attacks. Biotechnology and Bioengineering, 105(2), 221–230.
Hertz, E., Yu, A.C.H., Hertz, L., Juurlink, B.H.J., Schousboe, A., Preparation of primary cultures of mouse cortical neurons. In A Dissection and Tissue Culture Manual for the Nervous System, Shahar, A., De Vellis, J., Vernadakis, A., Haber, B., Ed. Alan R Liss: New York, 1989; pp 183–186.
Walls, A. B.; Bak, L. K.; Sonnewald, U.; Schousboe, A.; Waagepetersen, H. S., Metabolic mapping of astrocytes and neurons in culture using stable isotopes and gas chromatography-mass spectrometry (GC-MS). In Brain Energy Metabolism, Hirrlinger, J.; Waagepetersen, H. S., Eds. Springer New York: New York, NY, 2014; pp 73–105.
Tulpule, K.; Hohnholt, M. C.; Hirrlinger, J.; Dringen, R., Primary cultures of astrocytes and neurons as model systems to study the metabolism and metabolite export from brain cells #. In T Brain Energy Metabolism, Vol. 90, pp 45–72.
Drejer, J., Honore, T., & Schousboe, A. (1987). Excitatory amino acid-induced release of 3H-GABA from cultured mouse cerebral cortex interneurons. The Journal of Neuroscience, 7(9), 2910–2916.
Belhage, B., Hansen, G. H., & Schousboe, A. (1993). Depolarization by K+ and glutamate activates different neurotransmitter release mechanisms in gabaergic neurons: vesicular versus non-vesicular release of GABA. Neuroscience, 54(4), 1019–1034.
Mawhinney, T. P., Robinett, R. S. R., Atalay, A., & Madson, M. A. (1986). Gas-liquid chromatography and mass spectral analysis of mono-, di- and tricarboxylates as their tert.-butyldimethylsilyl derivatives. Journal of Chromatography A, 361, 117–130.
Biemann, K. (1962). Mass spectrometry: organic chemical applications. McGraw-Hill.
Mason, G. F., Rothman, D. L., Behar, K. L., & Shulman, R. G. (1992). NMR determination of the TCA cycle rate and alpha-ketoglutarate/glutamate exchange rate in rat brain. Journal of Cerebral Blood Flow and Metabolism, 12(3), 434–447.
Gram, L., Larsson, O. M., Johnsen, A. H., & Schousboe, A. (1988). Effects of valproate, vigabatrin and aminooxyacetic acid on release of endogenous and exogenous GABA from cultured neurons. Epilepsy Research, 2(2), 87–95.
Golan, H., & Grossman, Y. (1996). Block of glutamate decarboxylase decreases GABAergic inhibition at the crayfish synapses: possible role of presynaptic metabotropic mechanisms. Journal of Neurophysiology, 75(5), 2089–2098.
Golan, H., Talpalar, A. E., Schleifstein-Attias, D., & Grossman, Y. (1996). GABA metabolism controls inhibition efficacy in the mammalian CNS. Neuroscience Letters, 217(1), 25–28.
Waagepetersen, H. S., Sonnewald, U., Larsson, O. M., & Schousboe, A. (2000). Compartmentation of TCA cycle metabolism in cultured neocortical neurons revealed by 13C MR spectroscopy. Neurochemistry International, 36(4–5), 349–358.
Lapuente-Brun, E., Moreno-Loshuertos, R., Acin-Perez, R., Latorre-Pellicer, A., Colas, C., Balsa, E., Perales-Clemente, E., Quiros, P. M., Calvo, E., Rodriguez-Hernandez, M. A., Navas, P., Cruz, R., Carracedo, A., Lopez-Otin, C., Perez-Martos, A., Fernandez-Silva, P., Fernandez-Vizarra, E., & Enriquez, J. A. (2013). Supercomplex assembly determines electron flux in the mitochondrial electron transport chain. Science, 340(6140), 1567–1570.
Kann, O., & Kovacs, R. (2007). Mitochondria and neuronal activity. American Journal of Physiology. Cell Physiology, 292(2), C641–C657.
Gao, J.; Wang, L.; Liu, J.; Xie, F.; Su, B.; Wang, X., Abnormalities of mitochondrial dynamics in neurodegenerative diseases. Antioxidants (Basel) 2017, 6 (2).
Golpich, M., Amini, E., Mohamed, Z., Azman Ali, R., Mohamed Ibrahim, N., & Ahmadiani, A. (2017). Mitochondrial dysfunction and biogenesis in neurodegenerative diseases: pathogenesis and treatment. CNS Neuroscience & Therapeutics, 23(1), 5–22.
Acknowledgments
Heidi Nielsen is acknowledged for preparing the cell cultures.
Funding
This study was supported by grant no. 10-094362/0602-01660B from the Danish Medical Research Council (to Anne B. Walls).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of Interest
The authors declare that they have no competing interest.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
This work was performed in the laboratories of Winnie E. Svendsen and of Helle S. Waagepetersen
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Walls, A.B., Dimaki, M., Korsgaard, T. et al. Diphenylalanine Peptide Nanowires as a Substrate for Neural Cultures. BioNanoSci. 10, 224–234 (2020). https://doi.org/10.1007/s12668-019-00717-w
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12668-019-00717-w