Abstract
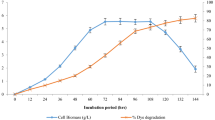
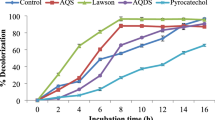
Alcaligenes species capable of degrading highly recalcitrant, carcinogenic, water-soluble dye—Congo red—were isolated from Indian West coastal sediments. Individual strains showed decolorization rates ranging from 76.49 to 98.76% within 24–48 h. Decolorization was most efficient at anoxic conditions catalyzed by intracellular azoreductase enzyme with an activity of 0.032 µmol min−1 mg−1 of protein. Degradation was confirmed by HPLC and FTIR analysis. LC/MS analysis of degraded metabolites established the cleavage of the azo bond-producing biphenyl diamine and 1,2′-diaminonapthalene-4-sulfonic acid. These results signify the effectiveness and ease to engineer processes such as feed batch/immobilized cell systems using these strains as biocatalysts to address the problem of global coastal water pollution caused by increased disposal of azo dye-containing industrial effluents.






Similar content being viewed by others
References
Akiyama T, Asfahl KL, Savin MC (2010) Broad-host-range plasmids in treated wastewater effluent and receiving streams. J Environ Qual 39:2211–2215
Anjaneya O, Souche SY, Santoshkumar M, Karegoudar TB (2011) Decolorization of sulfonated azo dye Metanil Yellow by newly isolated bacterial strains: Bacillus sp. strain AK1 and Lysinibacillus sp. strain AK2. J Hazard Mater 190:351–358
Babu SS, Mohandass C, Raj AV, Rajasabapathy R, Dhale MA (2013) Multiple approaches towards decolorization and reuse of a textile dye (VB-B) by a marine bacterium Shewanella decolorationis. Water Air Soil Pollut 224:1–12
Babu S, Mohandas C, Vijayaraj AS, Dhale MA (2015) Detoxification and color removal of Congo red by a novel Dietzia sp. (DTS26)—a microcosm approach. Ecotoxicol Environ Saf 114:52–60
Bauer AW, Kirby WMM, Sherris JC, Turck M (1966) Antibiotics susceptibility testing by a standardized single disk method. Am J Clin Pathol 45:493–496
Chakraborty S, Basak B, Dutta S, Bhunia B, Dey A (2013) Decolorization and biodegradation of Congo red dye by a novel white rot fungus Alternaria alternata CMERI F6. Bioresour Technol 147:662–666
Chen KC, Wu JY, Liou DJ, Hwang SCJ (2003) Decolorization of the textile dyes by newly isolated bacterial strains. J Biotechnol 101:57–68
CLSI (2005) Performance standards for antimicrobial susceptibility testing; 15th informational supplement. CLSI/NCCLS M 100-S 15. Clinical and Laboratory Standards Institute. Wayne, PA
Gopinath KP, Murugesan S, Abraham J, Muthukumar K (2009) Bacillus sp. mutant for improved biodegradation of Congo red: random mutagenesis approach. Bioresour Technol 100:6295–6300
Hiral S, Kumar JN, Patel K, Kumar RN (2015) Photocatalytic decoloration of three commercial dyes in the aqueous phase and industrial effluents using TiO2 nanoparticles. Desalination Water Treat. doi:10.1080/19443994.2015.1005147
Hu TL (1998) Degradation of azo dye RP2B by Pseudomonas luteola. Water Sci Technol 38:299–306
Jadhav UU, Dawkar VV, Ghodake GS, Govindwar SP (2008) Biodegradation of direct red 5B, a textile dye by newly isolated Comamonas sp. UVS. J Hazard Mater 158:507–516
Joshi T, Iyengar L, Singh K, Garg S (2008) Isolation, identification and application of novel bacterial consortium TJ-1 for the decolorization of structurally different azo dyes. Bioresour Technol 99:7115–7121
Kalyani DC, Telke AA, Dhanve RS, Jadhav JP (2009) Ecofriendly biodegradation and detoxification of reactive red 2 textile dye by newly isolated Pseudomonas sp. SUK1. J Hazard Mater 163:735–742
Kaushik P, Malik A (2015) Mycoremediation of synthetic dyes: an insight into the mechanism, process optimization and reactor design. microbial degradation of synthetic dyes in wastewaters. Environ Sci Eng. doi:10.1007/978-3-319-10942-8_1
Khan R, Bhawana P, Fulekar MH (2013) Microbial decolorization and degradation of synthetic dyes: a review. Rev Environ Sci Biotechnol 12:75–97
Kumar CG, Mongolla P (2015) Microbial degradation of basic dyes in wastewaters microbial degradation of synthetic dyes in wastewaters. In: Singh S (ed) Microbial degradation of synthetic dyes in wastewaters. Environmental science and engineering. Springer, Cham, pp 85–110
Kumar K, Devi SS, Krishnamurthi K, Dutta D, Chakrabarti T (2007) Decolorization and detoxification of Direct Blue 15 by a bacterial consortium. Bioresour Technol 98:3168–3171
Li X, Zhang J, Jiang Y, Hu M, Li S, Zhai Q (2013) Efficient biodecolorization/degradation of Congo red and Alizarin Yellow R by chloroperoxidase from Caldariomyces fumago: catalytic mechanism and degradation pathway. Ind Eng Chem Res 52:13572–13579. doi:10.1021/ie4007563
Mendes S, Pereira L, Batista C, Martins LO (2011) Molecular determinants of Azo reduction activity in the strain Pseudomonas putida MET94. Appl Microbiol Biotechnol 92:393–405
Morrison JM, Wright CM, John GH (2012) Identification, isolation and characterization of a novel azoreductase from Clostridium perfringens. Anaerobe 18:229–234
NCCLS (2002) Performance standards for antimicrobial disk susceptibility tests, 7th edn. In: Approved Standard National Committee for Clinical Laboratory Standards document M2-A7, NCCLS, Villanova, PA
Ng IS, Chen T, Lin R, Zhang X, Ni C, Sun D (2013) Decolorization of textile azo dye and Congo red by an isolated strain of the dissimilatory manganese-reducing bacterium Shewanella xiamenensis BC01. Appl Microbiol Biotechnol. doi:10.1007/s00253-013-5151-z
Phugare SS, Kalyani DC, Patil AV, Jadhav JP (2010) Textile dye degradation by bacterial consortium and subsequent toxicological analysis of dye and dye metabolites using cytotoxicity, genotoxicity and oxidative stress studies. J Hazard Mater 186:713–723
Robinson T, McMullan G, Marchant R, Nigam P (2001) Remediation of dyes in textile effluent: a critical review on current treatment technologies with a proposed alternative. Bioresour Technol 77:247–255
Romhanyi G (1971) Selective differentiation between amyloid and connective tissue structures based on the collagen specific topo-optical staining reaction with Congo red. Virchows Arch A 354:209–222
Saratale RG, Saratale GD, Kalyani DC, Chang JS, Govindwar SP (2009) Enhanced decolorization and biodegradation of textile azo dye Scarlet R by using developed microbial consortium-GR. Biores Technol 100(9):2493–2500
Saratale RG, Gandhi SS, Purankar MV, Kurade MB, Govindwar SP, Oh SE, Saratale GD (2013) Decolorization and detoxification of sulfonated azo dye CI Remazol red and textile effluent by isolated Lysinibacillus sp. RGS. J Biosci Bioeng 115:658–667
Talarposhti MA, Donnelly T, Anderson GK (2001) Colour removal from a simulated dye waste water using a two-phase anaerobic packed bed reactor. Water Res 35:425–432
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 30:2725–2729
Telke AA, Joshi SM, Jadhav SU, Tamboli DP, Govindwar SP (2010) Decolorization and detoxification of Congo red and textile industry effluent by an isolated bacterium Pseudomonas sp.SU-EBT. Biodegradation 21:283–296
Thakur JK, Paul S, Dureja P, Annapurna K, Padaria JC, Gopal M (2014) Degradation of sulphonated azo dye Red HE7B by Bacillus sp. and elucidation of degradative pathways. Curr Microbiol 69:183–191
Vijaykumar MH, Vaishampayan PA, Shouche YS, Karegoudar TB (2007) Decolorization of naphthalene-containing sulfonated azo dyes by Kerstersia sp. strain VKY1. Enzyme Microb Technol 40:204–211
Acknowledgements
The authors are grateful to Director CSIR-National Institute of Oceanography Goa and Scientist-in-charge RC—Mumbai—for providing the infrastructure facilities and financial support. The authors are also thankful to Dr. Supriya Shet Tilvi for FTIR analysis. The funding for this work was provided by Ministry of Earth Sciences (MOES), Government of India. This is the CSIR-NIO contribution number 6121.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Rights and permissions
About this article
Cite this article
D’Souza, E., Fulke, A.B., Mulani, N. et al. Decolorization of Congo red mediated by marine Alcaligenes species isolated from Indian West coast sediments. Environ Earth Sci 76, 721 (2017). https://doi.org/10.1007/s12665-017-7077-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s12665-017-7077-8