Abstract
The advances in biomedical imaging equipment have produced a massive amount of medical images that are generated by the different modalities. Consequently, a huge volume of data has been produced and caused a complex and time-consuming retrieving process of the relevant cases. To resolve this issue, the Content-Based Biomedical Image Retrieval (CBMIR) system is applied to retrieve the related images from the earlier patients’ databases. However, the previous handcrafted features methods that applied the CBMIR model have shown poor performance in many multimodal databases. In this paper, we focus on designing CBMIR technique using Deep Learning (DL) models. We present a new Multimodal Biomedical Image Retrieval and Classification (M-BMIRC) technique for retrieving and classifying the biomedical images from huge databases. The proposed M-BMIRC model involves three dissimilar processes as following: feature extraction, similarity measurement, and classification. It uses an ensemble of handcrafted features from Zernike Moments (ZM) and deep features from Deep Convolutional Neural Networks (DCNN) for feature extraction process. Additionally, the Hausdorff Distance based similarity measure is employed to identify the resemblance between the queried image and the images that exist in the database. Moreover, the classification process gets executed on the retrieval images using the Probabilistic Neural Network (PNN) model, which allocates the class labels of the tested images. Finally, the experimental studies are conducted using two benchmark medical datasets and the results ensure the superior performance of the proposed model in terms of different measures include Average Precision Rate (APR), Average Recall Rate (ARR), F-score, accuracy, and Computation Time (CT).
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
Recently, the progressive development of digital systems, multimedia, and memory space has produced a massive amount of images as well as multimedia contents. Medical and diagnostic works are highly beneficial from the advancements of digital storage and content processing. The clinics have diagnosed and examined the imaging services to generate a high quantity of clinical images’ details. Hence, the deployment of significant clinical Image Retrieval (IR) model is essential for physicians to browse the huge datasets. There are many of computation and management methods for handling and automating the clinical images. An effective method named CBMIR, has been applied for diagnosing numerous diseases which is considered as productive management systems that managing a large quantity of information. Without the existence of these systems, the processing and filtering of data are highly complicated.
The CBMIR methods focus on extracting text details like tags that required manual annotations which give a low efficiency and require more time, manual intervention, and medical expertise. There is an extremely needs for CBMIR methods to be applied in automatic classification and image retrieval. They can support the medical Decision Support Systems (DSS), and clinical works to identify the related details from huge repositories. Additionally, the Content-Based Image Retrieval (CBIR) is defined as a computer vision model which is applied to explore the related images in massive databases. This exploration depends upon the image properties such as color, texture, and structure which is derived from the image. The working principle of CBIR system is based on the determined features (Liu et al. 2007). Initially, the images are depicted with respect to their features that presented in high dimensional feature space. Followed by saving the affinity between images in the database and then, the Query Image (QI) has been estimated in a feature space under the application of distance measures like Euclidean distance. Therefore, the depiction of image data is accomplished by the image characteristics and the affinity measure which are considered as very important metrics. Kumar et al. (2019) presented a biomedical image retrieval system which uses Zernike moments (ZMs) for extracting features from CT and MRI medical images. But the presented model does not include the deep learning (DL) models into execution and it does not consider the highly correlated visual properties between various classes. Besides, several research works have applied PNN for image classification processes (Afify et al. 2020; Bodyanskiy et al. 2020; Agostini et al. 2020).
In order to develop a CBMIR method, the image classification is a major challenge because of the highly correlated visual properties between various classes, which intended in mitigated retrieval functions. To overcome this problem, the advanced Machine Learning (ML) methods have been applied for generating better classification. In past decades, important developments were established in ML and Artificial Intelligence (AI) along with Deep Learning (DL) approach. The major principle behind DL is the analogous to the performance of human brain where data is computed by several layers of transformation. Hence, DL approaches have implied better performance in CBIR domains. Additionally, DL modules have major involvement in different clinical applications like Brain Tumor (BT) prediction, blood flow quantification and visualization, Diabetic Retinopathy (DR), and massive number of cancer prediction domains. Therefore, there are high demands for improving the CBMIR operations because of the rapid development of clinical imaging model.
Moreover, the extensive dissemination of Picture Archiving and Communication Systems (PACS) in medical centers implied a progressive enhancement in the dimensions of medicinal clinical collections. Hence, managing the massive clinical databases required the deployment of efficient CBIR systems. Besides, managing the database, we required a particular CBMIR model which guides the physicians in making significant decisions regarding certain disease. By deriving the same images and histories, the physicians can make a critical decision regarding patient’s disorders and diagnoses. For clinical images, the global feature extraction depends upon failure of generating compact feature representations as medically significant details which are localized in tiny regions of image.
This paper presents a new Multimodal Biomedical Image Retrieval and Classification (M-BMIRC) technique to retrieve and classify the biomedical images from huge database. The proposed M-BMIRC model performs the retrieval and classification processes using feature extraction, similarity measurement, and classification. The presented model uses an ensemble of handcrafted features from ZM and deep features from DCNN) for feature extraction process. In addition to, the Hausdorff Distance based similarity measure which is employed to identify the resemblance between the QI and the images that exist in the database. Furthermore, the classification process is implemented on the retrieval images using the Probabilistic Neural Network (PNN) model, which allocates the class labels for the test images. The reset of this paper is organized as following: Sect. 2 presents the literature review for all related areas of the research. Followed by Sect. 3 which provides the details of the proposed model. Then, the experimental validation is presented in Sect. 4. Finally, the conclusion of this work is given in Sect. 5.
2 Literature survey
Numerous CBMIR models have been proposed in the literature. In this section, we present brief reviews of the two existing CBMIR models named handcrafted features using classical models and deep features using DL models.
2.1 Handcrafted features
For clinical images, the global feature extraction systems have ineffective compact feature implications in the tiny regions of image (Kumar and Gopal 2014). A method depends upon Bag of Visual Words (BoVWs) under the application of SIFT features has been projected for brain Magnetic Resonance Image (MRI) to diagnose Alzheimer disease (Mizotin 2012). Laguerre Circular Harmonic Functions coefficients (LG-CHF) has been employed as feature vectors for retrieving the surpassed Scale Invariant Feature Transform (SIFT), as well as Speeded up Robust Features (SURF) characteristics that relied on retrieving systems. Additionally, a model for CBIR for skin lesion images has been projected in (Jiji and Raj 2015) which provides a newly approach that depending on shape, color, and texture features of the image under processing using the regression trees. Maximum specificity and sensitivity have been addressed for skin lesion images.
An effective clinical CBIR system has been projected as a solution for doctor to learn mammographic images using various features extraction methods, diverse metrics, and relevance feedback (Ponciano-Silva et al. 2013). Two CBIR technologies according to wavelet adaptation based on discriminate wavelet have been applied to classify a QI that estimating the optimal wavelet filter in (Quellec et al. 2012). A regression performance undergoes tuning used to enhance the retrieving performance by employing various wavelet bases for QI. An essential improvement was attained in IR function. Moreover, a classification based supervised learning model applied to biomedical IR has been presented (Rahman et al. 2011). It has applied image extracting and affinity fusion depends upon multiclass Support Vector Machine (SVM) which is employed for predicting QI. Therefore, by reducing the imbalanced photographs, the search region is limited in massive databases for similarity estimation.
2.2 Deep learning-based models
Recently, an essential breakthrough in DL is performed in clinical applications which is categorized into 2 single modality-relied and multiple modalities-related methodologies of imaging. Initially, a single modality-relied technique and the two-phase CBMIR method have been employed for automated retrieval of radiographic images. Followed by the major class label that has been allocated under the application of CNN-relied features, and in second stage, the outlier images has been extracted from the detected class according to the low-level edge histogram features. Alternatively, CNN-relied model proposed in (Anthimopoulos et al. 2016) for classification of interstitial lung diseases (ILDs) patterns by filtration of ILD features from decided dataset. The traditional categorization of Restricted Boltzmann Machine (RBM)-relied approach developed to examining the lung CT scan by integrating both generative and discriminative representation learning (Van et al. 2016). A CNN-related automated classification of peri-fissural nodules (PFN) was introduced in (Ciompi et al. 2015) for performing the lung cancer scanning. Then, 2-stage multi-instance DL model has been projected for classifying diverse body organs (Yan et al. 2016). First, the CNN undergoes training on local patches used to isolate the differential and non-informative patches from the training samples. Second, the fine-tuned of filtered patches feed to the classification process.
A brief examination of DL in CAD introduced in (Shin et al. 2016). Hence, there are three major features like CNN structures, dataset scale, and the transfer learning of CNN. A fully automated 3D CNN model for detecting cerebral micro-bleeds (CMBs) from MRI (Dou et al. 2016). The CMBs are tiny hemorrhages (HM) blood vessels where the prediction offers a deep insight for cerebrovascular infections as well as cognitive dysfunctions. The effective CNN training technology has been projected by dynamic selection of negative instances at the training process which implies an optimal function in HM prediction inside a color fundus photographs. A multi-view convolutional network (ConvNets)-relied CAD model presented for predicting pulmonary lumps from lung computed tomography (CT) scan photographs (Setio et al. 2016). Since the multiple modalities-related models such as a DL-based technology for multiclass CBMIR resented in (Adnan et al. 2017) which classifies the multimodal clinical images. For example, the intermodal dataset which is composed of massive classes with modalities CT, MRI, fundus camera (Das et al. 2020), positron-emission tomography (PET), and optical projection tomography (OPT) has been applied for training the network.
3 The proposed model
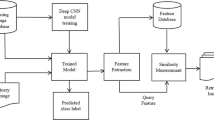
The development processes of the proposed CBMIR model is depicted in Fig. 1. Firstly, the collection of handcrafted features was extracted from ZM and the deep features were extracted using the Deep Convolutional Neural Networks (DCNN) from the input image. Using the new QI as input, a Hausdorff Distance based similarity measure is employed to determine the related images and retrieve them. Then, the PNN method is executed on the retrieved images to determine the actual class labels of them.
3.1 Feature extraction
In this section, we explain the integration of Zernike Moments (ZMs) with DCNN to extract the deep features. Initially, the ZMs are Zernike polynomial related to the orthogonal moments (Gau et al. 2018). The input image is feed into a tedious orthogonal Zernike polynomial to accomplish the ZMs. The major benefit of the ZM is deriving features by filtering texture and shape attributes to be comprised a minimum amount of repeated data. It is obtained in a diverse order of moments to depict the images with insensible towards the outliers, and it is a rotation invariant.
Hence, the ZMs across a unit disk is processed by:
where, \({Z}_{pq}\) implies the ZMs in conjunction with \(p\) order and \(q\) repetition for \(f(x, y),\pi (pi)\) is 3.14, \({V}_{pq}\) is Zernike orthogonal based function as well as \({V}_{pq}^{*}\) refers tedious conjugate of \({V}_{pq}\) which is estimated as:
where \(r=\sqrt{{a}^{2}-{b}^{2}},\Theta ={\mathrm{tan}}^{-1}\left(\frac{y}{\chi }\right),0\le \Theta \le 2\pi ,j=\sqrt{-1}\) and \({R}_{pq}(r)\) denotes a radial polynomial which is determined as:
The ZM functions for the 0th order approximation as well as external unit disk, which have been applied for enhancing the model performance. ZM coefficients \({f}_{ZM}\) are measured by assuming the minimum order of ZMs using Eq. (1). In order to extract the low order, the ZM features and the massive number of features have to be assumed in this process which refers to the metrics of \(p\) and \(q\) within \(0\) to 5, and the overall values of Zernike coefficient would be:
by applying Eq. (5)
where, \({Z}_{1},{Z}_{2},{Z}_{3},{Z}_{4},\dots {Z}_{m}\) means ZM coefficients and \(m\) defines the overall count of ZM coefficients.
where pmax means the higher value of \(p\).
Then, the deep features from DCNN model are employed. Researchers have identified that as a human visual system to resolve the issues that related to the classification, prediction, and identification, with robust deployment of significant nervous systems. This process promotes the biological visual systems to establish the latest data processing models. Cells in cortex of human optical system are suspicious for tiny regions and can approve where a local spatial correlation between cells in an image.
The CNN structure applies two specific models: the local receptive field and the distributed weights. An activation measure of a convolution neurons is estimated by enhancing the local input with weight \(W\), which has been distributed in an entire input space. The neurons which come under a similar layer share an identical weight. In general, CNN structure is composed of convolutional and pooling layers. Initially, convolution layer is embedded with a pooling layer, which reflects the features of tedious and simple cells in mammalian visual cortex [21]. The input data in CNN is a matrix with a \(3D\) spatial infrastructure, where \((H, W)\), (\({H}^{{\prime}}, W^{{\prime}}\)) and (\({H}^{{\prime\prime}}, {W}^{{\prime\prime}}\)) imply the size of spatial dimension of input data, convolution kernel, and output data respectively. The counts of convolution kernel feature vectors are implied by \(D\), and \({D}^{{\prime\prime}}\) that demonstrate the \(3D\) data, \(x\in {\mathbb{R}}^{H\times W\times D},\)
where \(x\) defines the input data, \(f\) implies a convolution filter, and \(y\) is the output data. The \(1D\) signal \(x\) undergoes the convolution by filter \(f\) to estimate the signal \(y\) as provided below:
where \({b}_{{d}^{{\prime\prime}}}\) refers to a neuron offset and \({f}_{{i}^{{\prime}}{j}^{{\prime}}d}\) indicates a convolution kernel matrix of dth \({i}^{{\prime}}\times {j}^{{\prime}}.\)
Once the convolution task is completed, the pooling layer is applied. The typical employed pooling task is the \(\mathrm{max}\) pooling. It is mainly applied to estimate the highest response of the feature channel in \({H}^{{\prime}}\times {W}^{{\prime}}\) region. A feature map is highly robust for data distortion and accomplishes a maximum invariance using pooling. Moreover, the pooling layer reduces the size of feature map and limits the processing overhead.
The classical CNN processing flow is developed with the data structure features for image classification tasks. Hence, training data is attained by using forward propagation to compute original output and the related actual real data tags competition. Stochastic Gradient Descent (SGD) has been applied for modifying the synapses and the attributes of a network structure, and various iterative trainings have been performed for developing the network system. The test data is projected as a network model. Hence, the final data is attained by feature extraction which are mapped with the original data, and the final outcomes are categorized using the competitive final model.
3.2 Similarity measurement
Feature extraction is defined as a significant step of CBMIR and the similarity estimation is an alternate key step in IR. Presently, the distance and the correlation measures are generally applied for measuring the similarity. When the distance among feature points of images are maximum, then the images are similar. Typically, the models can be Euclidean distance, Hausdorff distance, Manhattan distance, and EMD distance. Firstly, the Euclidean distance is a simple similarity estimation model (Xiaoming et al. 2018) that can be simple to learn. Additionally, it is sensible for image deformation. Secondly, the Manhattan distance depends upon the systems rotation of the coordinates while estimation; also the mapping can be applied instead of the coordinate rotation. Finally, the Hausdorff distance is not a corresponding to the distance from the various points, but it refers to the max–min distances. It comes under the fuzzy mapping among point sets that mimics the entire affinity of the clinical image’s features set. In this study, the Hausdorff distances between two clinical images are estimated. In case of query clinical image set, \(X=\{{x}_{1}, {x}_{2}, \dots , {x}_{N}\}\) as well as a specific image set \(Y=\{{y}_{1},{y}_{2}, \dots , {y}_{M}\}\) in clinical image database. Then, Eqs. (9), (10), and (11) are used to estimate the Hausdorff distance \(H(X,Y)\) of the similarity among two clinical images.
where
where \(\| \bullet \| \) means the distance from feature points \(x\) and \(y\) in the clinical images. The \(h(X,Y)\) shows the forward Hausdorf distance from point \(x\) to point \(y\). It estimates a distance regarding point \(\chi \) of set \(X\) of clinical image to be derived and point \(y\) of a particular image set \(Y\) in the clinical image database to accomplish the minimum distances. Using the points \(x\) in the attained clinical image set \(X\) and following the above steps, the point \(x\) gains a corresponding distance, then selects the maximum lower distances. \(h(Y, X)\) is named a backward Hausdorff distance, and the calculation process is similar to \(h(X,Y)\). Eventually, the highest point between \(h(X,Y)\) and \(h(Y, X)\) is selected as a Hausdorf distance of the queried clinical image and the specific image from the clinical image database.
From the above-defined distances, the Hausdorff distance between the queried clinical image and the special image from the database is determined. This distance is graded from minimum to maximum, and the images from a database correspond to the initial Hausdorff distances are selected for retrieving the simulation outcome.
3.3 Image classification
In this stage, every retrieved image is classified using PNN. Basically, the PNN is significantly applied for the ship discovery, noise categorization and image classification. It is defined as a nonlinear classifier and the parallel model depends upon Bayesian lower risk criterion (Liao et al. 2018). The sample that has to be found is \(x\), and the posterior possibility \(P({S}_{k}|x)\) could be accomplished using PNN classifier. Therefore, when the possible densities of the classes are isolated, the training instances are learned, and the purpose of the identified class is determined. Consequently, the trained PNN has been applied for computing the identity of \(x\). A general PNN classification method is composed to the input layer, pattern layer, summation layer and output layer. The Architecture design of PNN is illustrated in Fig. 2. Initially, the input layer neuron have been applied to gain measures from the training instances and forwards the data to neurons from the pattern layer, that is completely associated with the input layer. Then, assign the count of the neurons in the input layer to the length of the input vector. Additionally, the count of neurons that present to the pattern layer is equal to the count of training samples. In this approach, neurons are gathered into various groups, and ith neuron grouped in \(k\) considers as a Gaussian function \({f}_{i}^{(k)}(x, \sigma ),i=\mathrm{1,2}, \cdots , {m}_{k}\), where \({m}_{k}\) implies the count of the neurons in group \(k,k=\mathrm{1,2}, \cdots ,K\). Moreover, the Gaussian function is named as a probability density function which is demonstrated in the following:
where \(\nu \) means the dimension of the input vector \(x=({x}_{1},{x}_{2}, \cdots , {x}_{v}),{x}_{j}\) that implies the jth component of the input vector \(x, {x}_{ij}^{(k)}\) shows the jth unit of ith neuron in class \(k\).
Thus, it is named as smoothing attribute \(\sigma \in (\mathrm{0,1})\) that estimated experimentally by relating the corresponding classification accuracy which is significant factor in calculating the error of PNN classification approach. Finally, the outputs of pattern layer is linked with the summation units which depends upon class of the patterns. The neurons for all groups and a neuron in summation layer add to the results that gained from pattern layer neurons as given below:
Lastly, the final layer neuron provides value of 1 and various measures \(0.e\) value of 1 determines the classifier’s solution for input vectors. Especially, the input vector \(\chi \) comes under the class \(k\) when \({p}_{k}(x)>{p}_{{k}^{{\prime}}}(x)\) for all \({k}^{{\prime}}=\mathrm{1,2},\cdots ,K\) and \(k\ne {k}^{{\prime}}.\)
Therefore, the key objective of PNN training is identifying the best estimates of probability density function based on the training instances as well as the labels to assure the classifier under a state of the lower error rate as well as the threats. If the samples examined and forwarded to the pattern layer, then the result of a neuron is measured on the basis of the trained density function. Also, the identified outcomes are attained by various computations in summation and output layers. We gain the following benefits when apply the PNN as a classifier for signals classification:
-
1.
It composes a simple architecture, and it is simple for training. In PNN-related probability density function evaluation, the weight of the neuron in pattern is consumed from an input sample value.
-
2.
The training process of a network is more sophisticated, and there is no need for retraining prolonged duration in both inclusion and deletion of the groups.
-
3.
It is impossible to generate local optimal solutions, and the precision is maximum when compared with alternate classifiers. Additionally, the complexity of classification issues is sufficient for training instances, and the best solution can be accomplished by Bayes criterion.
Algorithm 1: Pseudo-code of PNN | |
|---|---|
1. Initialize PNN (Train set, Test set, Spread) 2. Provide train set and class details 3. Estimate \({N}_{i},P(C)\) where \({N}_{i}\)‐No. of training patterns in class \({C}_{i}\); 4. Initialize PNN spread as \(0=0.25\) and fix counter \(=1.\) 5. Select and observation of \({X}_{test}\) from test set 6. Do counter \(=\) counter \(+\) 1 7. Estimate unconditional probability \(p({X}_{test})\) as well as conditional probability \(P({X}_{test}/{C}_{i})\) 8. Estimate posterior probability of \({X}_{test}\) 9. Estimate the maximum count of inputs from pattern units 10. Until counter \(=M\) 11. The categorization of a pattern vector is developed on the basis of Bayesian Rule 12. End Procedure | |
4 Experimental validation
The performance of the M-BMIRC model is validated against two benchmark datasets NEMA CT (Shahid et al. 2022) and OASIS MRI (Siddiqu et al. 2022). The former NEMA CT database comprises a set of 600 images with a resolution of 512 × 512 pixels and the dataset comprises the images under ten distinct class labels. Additionally, the OASIS MRI database consists of a collection of 416 MRI images with the size of 208*208 pixels and the dataset holds images under four different class labels. Few set of sample test images are illustrated in Fig. 3 and the details are provided in Table 1. The parameter setting of the DCNN model is given as follows: learning rate: 0.05, batch size: 500, maximum epoch count: 15, dropout rate: 0.2, and momentum: 0.9.
Table 2 and Fig. 4 present the detailed comparative retrieval results analysis of the M-BMIRC model on the applied NEMA CT images. The experimental results denoted that the LNIP model has shown insufficient retrieval performance by obtaining a minimum ARR of 0.1624, APR of 0.0867, and F-score of 0.1131. Next, the LNDP model has shown noticeably better retrieval results over the LNIP model with the ARR of 0.1642, APR of 0.1009, and F-score of 0.125. Followed by, the LBP model that has depicted slightly higher retrieval results with the ARR of 0.1659, APR of 0.0879, and F-score of 0.1149. Also, the LTDP model has demonstrated moderate results with the ARR of 0.1659, APR of 0.0897, and F-score of 0.1164. Furthermore, the DLTerQEP model has shown manageable results with the ARR of 0.1668, APR of 0.091, and F-score of 0.1177. Moreover, the LDGP model has tried to demonstrate better results over the earlier models with the ARR of 0.1728, APR of 0.0905, and F-score of 0.1188. Accordingly, the LDEP model has attained somewhat higher results with the ARR of 0.1839, APR of 0.1027, and F-score of 0.1318. Eventually, the LBDISP model has resulted a manageable result with the ARR of 0.1905, APR of 0.1011, and F-score of 0.1321. Moreover, the LWP and LBDP models have achieved near identical and slightly satisfactory results over the other methods except OFMMs and M-BMIRC model. Though the OFMMs model has produced near optimal results with the ARR of 0.7412, APR of 0.4851, and F-score of 0.5864. Finally, the proposed M-BMIRC model has shown superior results with the ARR of 0.7853, APR of 0.6285, and F-score of 0.7309.
Table 3 and Fig. 5 present the detailed comparative retrieval results analysis of the M-BMIRC model in the terms of ARR, APR, and F-score. The experimental outcomes denoted that the LNDP model has shown insufficient retrieval performance by obtaining a minimum ARR of 0.2378, APR of 0.2499, and F-score of 0.2437. Next, the DLTerQEP model has shown noticeably optimal retrieval results over the LNDP model with the ARR of 0.2395, APR of 0.2550, and F-score of 0.247. Followed by, the LBP model that has depicted slightly higher retrieval results with the ARR of 0.2401, APR of 0.2559, and F-score of 0.2478. Also, the LNIP model has presented moderate results with the ARR of 0.2401, APR of 0.2655, and F-score of 0.2521. Furthermore, the LDEP model has shown manageable results with the ARR of 0.2403, APR of 0.2389, and F-score of 0.2396. Moreover, the LTDP model has demonstrated better results over the earlier models with the ARR of 0.2406, APR of 0.2606, and F-score of 0.2502. Likewise, the LDGP model has presented even better results with ARR of 0.2411, APR of 0.2660, and F-score of 0.253. Accordingly, the LBDISP model has attained superior results with the ARR of 0.2431, APR of 0.2472, and F-score of 0.2451.
Eventually, the LWP model has resulted manageable results with the ARR of 0.2447, APR of 0.2556, and F-score of 0.2500. Moreover, the LBDP models have achieved near slightly satisfactory results with the ARR of 0.2497, APR of 0.2428, and F-score of 0.2462. However the OFMMs model has presented near optimal results with the ARR of 0.3039, APR of 0.3376, and F-score of 0.3198. Finally, the proposed M-BMIRC model has shown superior results with the ARR of 0.4298, APR of 0.4467, and F-score of 0.4343.
Table 4 and Figs. 6 and 7 present detailed comparative retrieval outcomes analysis of the M-BMIRC model using NEMA CT and OASIS MRI images. The experimental outcomes denoted that the LNIP model has exhibited insufficient retrieval performance by obtaining a highest CT of 497.72 s and 66.92 s when applied on NEMA CT and OASIS MRI images. Next, the DLTerQEP model has demonstrated noticeably optimal retrieval results over the LNIP model with the CT of 481.54 s and 154.84 s when applied on NEMA CT and OASIS MRI images.
Followed by, the LTDP model that has depicted somewhat better retrieval outcomes with the CT of 332.75 s and 49.64 s when applied on NEMA CT and OASIS MRI images. Likewise, the LBDISP model has exhibited moderate outcomes with the CT of 276.58 s and 36.34 s when applied on NEMA CT and OASIS MRI images. Furthermore, the LBDP model has shown somewhat manageable results with the CT of 183.39 s and 25.68 s when applied on NEMA CT and OASIS MRI images. Moreover, the LDEP model has demonstrated better outcomes over the earlier models with the CT of 157.99 s and 18.97 s when applied on NEMA CT and OASIS MRI images. Accordingly, the LNDP model has attained slightly lower CT of 157.94 s and 22.77 s when applied on NEMA CT and OASIS MRI images. Eventually, the LWP model has produced a manageable result with the CT of 109.76 s and 17.25 s when applied on NEMA CT and OASIS MRI images. Moreover, the LWP and OFMMs models have achieved near same and slightly satisfactory results over the other methods except LDGP and M-BMIRC model.
Although, the DGP model has displayed near optimal results with the CT of 55.71 s and 7.9 s when applied on NEMA CT and OASIS MRI images. Finally, the proposed M-BMIRC model has shown superior results with the minimum CT of 51.82 s and 6.18 s when applied on NEMA CT and OASIS MRI images.
Table 5 and Fig. 8 present detailed comparative retrieval results analysis of the M-BMIRC model in terms of classification accuracy. The experimental results denoted that the SVM model has shown insufficient retrieval performance by obtaining maximum accuracy equals to 76.36% and 78.38% when applied on NEMA CT and OASIS MRI images. Next, the MLP model has shown noticeably better retrieval results over the LNIP model with the accuracy 77.41% and 80.34% when applied on NEMA CT and OASIS MRI images. Followed by the ELM model that has depicted slightly better retrieval results with the accuracy equals to 79.37% and 81.78% when applied on NEMA CT and OASIS MRI images. Similarly, the KSVM model has exhibited moderate results with the accuracy of 80.06% and 80.14% when applied on NEMA CT and OASIS MRI images. Furthermore, the KELM model has shown somewhat manageable results with the accuracy of 80.12% and 82.86% when applied on NEMA CT and OASIS MRI images. But, the presented PNN model has showcased superior results with the minimum accuracy equals 87.45% and 89.08% when applied on NEMA CT and OASIS MRI images.
5 Conclusion
This paper has developed a new M-BMIRC technique for retrieving and classifying the biomedical images from huge databases. The proposed model performs the retrieval and classification processes using feature extraction, similarity measurement, and classification. Firstly, the ensemble of handcrafted features using ZM and deep features from DCNN is extracted from the input image. On the application of QI as input, the Hausdorff Distance based similarity measure is employed to determine the related images to be retrieved. Finally, the PNN model is applied to the retrieved images to determine the actual class labels of them. For experimental validation, two benchmark medical datasets are used, and the experimental results ensured the superior performance of the proposed model over the existing methods. In future, the outcome of the presented model can be enhanced using the learning rate scheduling technique for DCNN.
References
Afify M, Mohammed K, Hassanien A (2020) Multi-images recognition of breast cancer histopathological via probabilistic neural network approach. J Syst Manag Sci 1(2):53–68
Adnan Q, Anwar S, Awais M, Majid M (2017) Medical image retrieval using deep convolutional neural network. Neurocomputing 266:8–20
Agostini C, Sampaio M (2020) Probabilistic neural network with Bayesian-based, spectral torque imaging and deep convolutional autoencoder for PDC bit wear monitoring. J Petrol Sci Eng 193:107434
Anthimopoulos M, Christodoulidis S, Ebner L, Christe A, Mougiakakou S (2016) Lung pattern classification for interstitial lung diseases using a deep convolutional neural network. IEEE Trans Med Imaging 35:1207–1216
Bodyanskiy Y, Deineko A, Pliss I, Chala O, Nortsova A (2020) Matrix fuzzy-probabilistic neural network in image recognition task. In: IEEE third international conference on data stream mining & processing (DSMP), Los Angeles, pp 33–36
Ciompi F, Hoop B, Riel V, Chung K, Scholten E, Oudkerk M, Jong P (2015) Automatic classification of pulmonary peri-fissural nodules in computed tomography using an ensemble of 2D views and a convolutional neural network out-of-the-box. Med Image Anal 26:195–202
Das P, Neelima A (2020) A robust feature descriptor for biomedical image retrieval. IRBM 42(4):245–257
Dou Q, Chen H, Yu L, Zhao L, Qin J, Wang D, Mok V, Shi L, Heng P (2016) Automatic detection of cerebral microbleeds from MR images via 3D convolutional neural networks. IEEE Trans Med Imaging 35:1182–1195
Gao H, Lin S, Yang Y, Li C, Yang M (2018) Convolution neural network based on two-dimensional spectrum for hyperspectral image classification. J Sens 8602103:1–12
Jiji G, Raj P (2015) Content-based image retrieval in dermatology using intelligent technique. IET Image Process 9(4):306–317
Kumar K, Gopal T (2014) A novel approach to self order feature reweighting in CBIR to reduce semantic gap using relevance feedback. In: Proceedings of the international conference on circuit, power and computing technologies (ICCPCT), Bhopal, pp 1437–1442
Kumar Y, Aggarwal A, Tiwari S, Singh K (2018) An efficient and robust approach for biomedical image retrieval using Zernike moments. Biomed Signal Process Control 39:459–473
Liao X, Li B, Yang B (2018) A novel classification and identification scheme of emitter signals based on ward’s clustering and probabilistic neural networks with correlation analysis. Comput Intell Neurosci 1458962:1–14
Liu Y, Zhang D, Lu G, Ma Y (2007) A survey of content-based image retrieval with high-level semantics. Pattern Recogn 40(1):262–282
Mizotin M, Benois-Pineau J, Allard M, Catheline G (2012) Feature-based brain MRI retrieval for Alzheimer disease diagnosis. In: Proceedings of the ninteenth IEEE international conference on image processing, Valencia, pp 1241–1244
Ponciano-Silva M, Souza J, Bugatti P (2013) Does a CBIR system really impact decisions of physicians in a clinical environment? In: Proceedings of the twenty-sixth IEEE international symposium on computer-based medical systems, Portugal, pp 41–46
Quellec G, Lamard M, Cazuguel G, Cochener B, Roux C (2012) Fast wavelet-based image characterization for highly adaptive image retrieval. IEEE Trans Image Process 21(4):1613–1623
Rahman M, Antani S, Thoma S (2011) A learning-based similarity fusion and filtering approach for biomedical image retrieval using SVM classification and relevance feedback. IEEE Trans Inf Technol Biomed 15(4):640–646
Setio A, Ciompi F, Litjens G, Gerke P, Jacobs C, Riel V (2016) Pulmonary nodule detection in CT images: false positive reduction using multi-view convolutional networks. IEEE Trans Med Imaging 35:1160–1169
Shahid A, Bazargani M, Banahan P, Namee B (2022) A two-stage de-identification process for privacy-preserving medical image analysis. Healthcare 10(5):1–18
Shin C, Roth H, Gao M, Lu L, Xu Z, Nogues I, Yao J (2016) Deep convolutional neural networks for computer-aided detection: CNN architectures, dataset characteristics and transfer learning. IEEE Trans Med Imaging 35:1285–1298
Siddiqi M, Azad M, Alhwaiti Y (2022) An enhanced machine learning approach for brain MRI classification. Diagnostics 12(11):1–16
Van T, Bruijne M (2016) Combining generative and discriminative representation learning for lung CT analysis with convolutional restricted Boltzmann machines. IEEE Trans Med Imaging 35:1262–1272
Xiaoming S, Ning Z, Haibin W, Xiaoyang Y, Xue W, Shuang Y (2018) Medical image retrieval approach by texture features fusion based on hausdorff distance. Math Probl Eng 7308328:1–11
Yan Z, Zhan Y, Peng Z, Liao S, Shinagawa Y, Zhang S (2016) Multi-instance deep learning: discover discriminative local anatomies for bodypart recognition. IEEE Trans Med Imaging 35:1332–1343
Funding
Open access funding provided by The Science, Technology & Innovation Funding Authority (STDF) in cooperation with The Egyptian Knowledge Bank (EKB).
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Mansour, R.F. Multimodal biomedical image retrieval and indexing system using handcrafted with deep convolution neural network features. J Ambient Intell Human Comput 14, 4551–4560 (2023). https://doi.org/10.1007/s12652-023-04575-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12652-023-04575-z