Abstract
Detecting COVID-19 from medical images is a challenging task that has excited scientists around the world. COVID-19 started in China in 2019, and it is still spreading even now. Chest X-ray and Computed Tomography (CT) scan are the most important imaging techniques for diagnosing COVID-19. All researchers are looking for effective solutions and fast treatment methods for this epidemic. To reduce the need for medical experts, fast and accurate automated detection techniques are introduced. Deep learning convolution neural network (DL-CNN) technologies are showing remarkable results for detecting cases of COVID-19. In this paper, deep feature concatenation (DFC) mechanism is utilized in two different ways. In the first one, DFC links deep features extracted from X-ray and CT scan using a simple proposed CNN. The other way depends on DFC to combine features extracted from either X-ray or CT scan using the proposed CNN architecture and two modern pre-trained CNNs: ResNet and GoogleNet. The DFC mechanism is applied to form a definitive classification descriptor. The proposed CNN architecture consists of three deep layers to overcome the problem of large time consumption. For each image type, the proposed CNN performance is studied using different optimization algorithms and different values for the maximum number of epochs, the learning rate (LR), and mini-batch (M-B) size. Experiments have demonstrated the superiority of the proposed approach compared to other modern and state-of-the-art methodologies in terms of accuracy, precision, recall and f_score.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
The novel COVID-19 disease has appeared in China in Wuhan city (Zhu et al. 2020). Then, it has become a pandemic, and it has rapidly spread through the world. It has had destructive effects on public health, daily life, and global economy (Singhal 2020). The symptoms of this disease include cough, sore throat, runny nose, fever, headache, and lung infection (Huang et al. 2020). Pneumonia is one of the main effects of COVID-19, which must be promptly diagnosed. Thus, rapid diagnostic testing of COVID-19 has become a major challenge. The laboratory-testing Real Time Polymerase Chain Reaction (RT-PCR) is the most famous current diagnostic tool. However, it is expensive (in some countries), and time-consuming (Xie et al. 2020a). Nowadays, medical image processing has a vital and influential role in diagnosing and detecting the disease (Xie et al. 2020b; Narin et al. 2020). Convolutional Neural Networks (CNNs) can be beneficial for accurate classification of this disease, and they can also help to overcome the shortage of specialists, especially in villages. In contrary to traditional learning paradigms, transfer learning seeks to solve the related problems by using the pre-gained knowledge (Jin et al. 2020; Najafabadi et al. 2015; Chen and Lin 2014; Singh et al. 2017).
Concatenation of deep features extracted from multiple CNNs produces a discriminant and appropriate descriptor for the classification process. Noreen et al. (2020) designed a feature concatenation algorithm for brain tumor classification. In this algorithm, two pre-trained models (Inception-v3 and DensNet201) have been used for deep feature extraction. DU. Chen et al. (2020) proposed a Selective Feature Connection Mechanism (SFCM), by which low-level and high-level features are linked together with a feature selector. Their mechanism can effectively demonstrate the effectiveness in multiple computer vision tasks including object detection, image classification, scene text detection, and image-to-image translation. Furthermore, Chenhui et al. Ma et al. (2019) presented a CNN-based fusion technique to integrate feature maps from various layers.
In this paper, we propose a DFC-based approach for automatic identification of COVID-19. This approach is applied to form a definitive classification descriptor. It is adopted in two different methods. The first method depends on the concatenation of deep features that are extracted separately from X-ray and CT images. Deep features are obtained using a proposed simple CNN that includes three deep layers, only. The second method aims to reach the optimal performance from either X-ray or CT scan by performing concatenation between the features extracted using three different CNNs, which are the proposed CNN architecture and two modern pre-trained CNNs (ResNet and GoogleNet). The main contributions of this paper can be summarized as follows:
-
Providing a CNN-based framework for diagnosis and detection of COVID-19 disease.
-
Introducing a simple proposed CNN architecture to solve the time consumption problem.
-
Adjusting the CNN parameters such as optimization algorithm, maximum number of epochs, LR, and M-B size.
-
Performing a DFC mechanism to take the advantage of combining deep features extracted from different image types.
-
Executing extensive experiments to compare the performance of the proposed approach with related ones.
The rest of the paper is organized as following. Section 2 describes the related work. Section 3 provides the proposed DFC mechanism. Section 4 shows the experimental results and discussions. Finally, the conclusion is summarized in Sect. 5.
2 Related work
The CNN is a class of deep learning tools. There are CNN models trained on instance ImageNet and used for image classification (Krizhevsky et al. 2012). The AlexNet consists of eight layers. It was proposed in 2012 (Jmour et al. 2018a). Furthermore, in 2014, VGGNet was developed to have different numbers of layers (11, 16 and 19 layers) Sengupta et al. (2019). In addition, LeNet was announced with a structure of two convolution layers, two max-pooling layers, a fully-connected layer and a classification layer (LeCun et al. 2015). Furthermore, GoogleNet, developed by Christian in 2014, had 22 layers with the insertion of inception layers (Szegedy et al. 2015a). Besides, ResNet, proposed by Kaiming in 2015, consisted of different layers (18, 50, 101, 152, and 1202 layers) He et al. (2016). Furthermore, SqueezeNet, presented in 2016, provided less parameters with faster performance than that of the AlexNet Iandola et al. (2016). In 2017, DenseNet introduced dense connection between layers (Huang et al. 2017). In 2018, MobileNetV2 was presented using the idea of inverted residual structures, and it performed well on mobile devices (Sandler et al. 2018). In addition, an efficient network was introduced in 2019 that includes 5.3 million parameters (Tan and Le 2019).
Nowadays, CNNs accomplished a great success in the areas of medical image analysis such as brain tumor detection (Sajjad et al. 2019), classification of skin diseases (Esteva et al. 2017), diagnosis of pulmonary modules (Cheng et al. 2016), diagnosis of breast cancer (Braman et al. 2019) and recognition of immune cells (Chen and ChefdHotel 2014). Recently, additional medical applications appeared such as improving the storage and exchange of DICOM medical data on the cloud (Dorgham et al. 2018; Zheng et al. 2017), transformation of secret information by using virtue of quantum-watermarked images Qu et al. (2019), Nuclei segmentation of brain tumors Guo et al. (2018), and early detection of COVID-19 disease. Furthermore, a healthcare framework was developed through a medical image forgery detection system to ensure that images are related to the healthcare system or not (Ghoneim et al. 2018; Amato et al. 2018).
A series of CNN-based approaches were designed for COVID-19 detection. Ying et al. and Song et al. (2020) used ResNet-50 for COVID-19 detection from CT images. Images were divided into 101 bacterial pneumonia, 86 normal cases and 88 COVID-19 patients. Their methodology achieved an accuracy of 86.0% for classification of bacterial or COVID-19 pneumonia. Besides, Farooq and Hafeez (2020) adopted ResNet-50 and applied various augmentation techniques to achieve an accuracy of 96.23%. Moreover, Xu et al. (2020) used V-Net for infected region segmentation. Also, ResNet-18 was used for diagnosis of COVID-19 cases with an accuracy of 86.7%. Hall et al. (2020) utilized the VGG16 model with 10-folds for training with the entire dataset to reach an accuracy of 96.1% and an Area Under the Curve (AUC) of 99.7%.
Nguyen et al. (2018) used ResNet-152, Inception-v3 and Inception-ResNet-v2 for extracting deep features from input images. These features are combined to represent all details of the input PAP-Smear and 2D-Hela datasets. Their mechanism provided an accuracy of 92.6%. Liang et al. (2019) studied the fusion of both CT and MR images for training of their Multi-layer Concatenation Fusion Network (MCFNet). They achieved good classification results. Rahimzadeh and Attar (2020) proposed a concatenation-based approach to link the features extracted using ResNet-50V2 and Xception CNNs. Their approach was applied on an X-ray dataset that was divided into three categories: pneumonia, normal and COVID-19 images. An accuracy of 91.4% was achieved.
3 The proposed DFC methodology
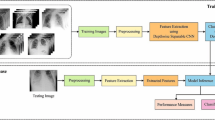
As mentioned before, the proposed methodology aims at accurate diagnosis of COVID-19 disease through performing concatenation between deep features extracted from X-ray and CT images. Initially, the input images are prepared appropriately through performing the image segmentation process to extract the Region Of Interest (ROI). Then, CNNs are used for the deep feature extraction process. Deep features are extracted in two different methods. For the first one, a simple proposed CNN extracts features from X-ray and CT images as presented in Fig. 1. In the other method, three CNNs (the proposed CNN, ResNet-18, and GoogleNet) extract features from either X-ray or CT images as shown in Fig. 2. Finally, the DFC methodology is applied to combine the extracted features into a single descriptor for classification. Some confirmed COVID-19 and non-COVID-19 images are given in Figs. 3 and 4 for CT and X-ray images, respectively.
3.1 Image segmentation
The image segmentation process is utilized to separate the pixels of interest and to locate the active contours of the lungs in input images (Michailovich et al. 2007). It can be performed with a snake in its intuitive manner, as it can be utilized to track objects. The contour is like a parametric curve \(V(s)=\left\{ X(s), Y(s)\right\}\), where \(X\left( s\right)\), and \(Y\left( s\right)\) are coordinate functions in a parametric domain “s”. Active contour could be represented using an energy function as follows (Leymarie and Levine 1993):
where \(E_{internal}^i\) and \(E_{external}^i\) are the internal and external energies at the \(i^{th}\) position of the contour. The internal energy represents the contour strength as:
where \(\alpha\) is the parameter that controls the snake tension and \(\beta\) is the rigidity parameter. The external energy is responsible for moving the snake to the borders or ROI in an image. The external energy could be computed as:
Figure 5 depicts examples of CT and X-ray images before and after performing the segmentation process.
3.2 Deep feature extraction
The process of feature extraction from images after segmentation is performed using CNNs. The CNNs are deep learning algorithms that contain multiple layers that are used for feature detection. These layers are convolutional, pooling, batch normalization, Rectified Linear Unit (ReLU), SoftMax, and fully-connected layers (Jmour et al. 2018b; Szegedy et al. 2015b). The convolutional layer depends on a number of kernels of weights \(\textbf{w}^l\) for each layer l to extract the local features from the input \(\textbf{x}^{l-1}\) as described in the following equation:
where \(\textbf{C}^l\) is the output feature map obtained by computing the dot products between the kernels and the input with added bias \({b}^l\).
The pooling layer has two essential types: maximum and average pooling. The output of the pooling process \(P^{l}\) is the down-sampled version of the entire feature map \(\textbf{C}^{l}\) depending on the window size (m, n) as:
The fully-connected layer is the last essential layer. Let the layer l-1 be the fully-connected layer. Layer l expects \(m_1^{(l-1)}\) feature maps having a size of \(m_2^{(l-1)}*m_3^{(l-1)}\) as input. The output of a fully-connected layer \(\textbf{Y}_i^{(l)}\) could be described as:
where \(\textbf{w}_{i,j,r,s}^{(l)}\) represents the weights used to connect the position (r, s) in the \(i^{th}\) unit in layer l and the jth feature map of layer \(l-1\). Here, we propose a new CNN model that includes three deep layers only to avoid issues such as complexity and large time consumption. Table 1 provides the architecture of the proposed model in detail.
3.3 Deep feature concatenation (DFC)
Feature concatenation is an effective way to add different features together to enhance the classification process. In this work, the DFC is applied in two different ways. In the first one, CT and X-ray features are extracted using the proposed CNN. Then, these features are linked to form the classification descriptor as:
On the other hand, the second way aims to obtain remarkable results from each dataset (CT or X-ray), separately. This can be accomplished through extracting features from images using three different CNNs: the proposed, ResNet-18, and GoogleNet. These features are combined together using the DFC as follows:
3.4 Classification
The classification process depends on the final feature descriptor to determine whether the input image is positive (COVID-19) or negative (Non-COVID-19). The fully-connected layer converts the input data into 1D vector. Then, the softmax layer calculates the scores of the classes as:
where \(\textbf{f}_{i}\) denotes features and \(y_{i}\) is the true class label of the \(i^{th}\) image. \(\textbf{w}_{j}\) and \(b_{j}\) are the weights and bias of the \(j^{th}\) class, respectively. N is the number of training samples and K is the number of classes.
4 Results and discussions
In this section, we discuss the results of the proposed DFC approach that could be implemented in two different methods. X-ray and CT image datasets are chosen to evaluate the proposed methodology (El-Shafai and El-Samie 2020). Each dataset includes positive (COVID) cases (2628 images) and negative (Non-COVID) cases (1620 images). For each class, images are divided into 70% for training and 30% for testing. Furthermore, the system performance is measured in terms of accuracy, precision, recall, and fscore. As discussed before, a new CNN model is proposed. In order to achieve its optimal performance, its hyper-parameters are adjusted including the optimization algorithm (Adam, RMS Prop, or SGDM), maximum number of epochs (30, 40, or 50), learning rate (LR) (0.001 or 0.0001), and Mini-Batch (MB) size (32, 64, or 128). Tables 2, 3, and 4 show the performance of the proposed model for the CT image dataset using a maximum number of epochs of 30, 40, and 50, respectively. For X-ray images, Tables 5, 6, and 7 present the results of the proposed CNN using maximum number of epochs of 30, 40, and 50, respectively. For each case, Figs. 6, 8, 10, 12, 14, and 16 provide accuracy versus iterations and loss versus iterations. Besides, for each case, Figs. 7, 9, 11, 13, 15, and 17 depict a graphical representation of the performance of the proposed model. In addition, the mean, variance, and standard deviation are also calculated.
From the previous results, it can be deduced that to reach the optimal performance of the proposed model, the proposed CNN hyper-parameters are adjusted as follows:
-
For CT images: Rmsprop algorithm for optimization, maximum number of epochs = 50, MB size = 64 and LR = 0.0001.
-
For X-ray images: Rmsprop algorithm for optimization, maximum number of epochs = 50, MB size = 32 and LR = 0.001.
These variables are adjusted with the mentioned values for each dataset during the implementation of the proposed approach. Now, the experimental results of the proposed approach (first method) will be discussed. Table 8 illustrates a comparison between the utilization of the proposed CNN to classify each dataset separately and the application of the proposed approach (first method) that combines both X-ray and CT features using DFC technique. The results in Table 8 confirm the superiority of the proposed approach and the improvement of the classification process as a result of the utilization of the DFC methodology. Additionally, Figure 18 shows a graphical comparison between the proposed CNN (CT Dataset), the proposed CNN (X-ray dataset), and the proposed approach (first method). In addition, Figures 19 and 20 present the ROC curves that demonstrate the superiority of the proposed DFC approach.
The main purpose of the second method of the proposed DFC approach is to obtain distinctive results from each image dataset X-ray or CT by performing concatenation between the features extracted from the dataset. Features are extracted using three different CNNs, which are the proposed CNN, ResNet-18, and GoogleNet. The results of the proposed approach (second method) are compared with the results of single-stage methods (single stage (ResNet-18), single-stage (GoogleNet), and single-stage (Proposed CNN)). Tables 9 and 10 display these results for both X-ray and CT datasets, respectively. By analyzing the results in Tables 9 and 10, it is clear that the proposed approach (second method) is significantly superior to the single-stage methods for both datasets. This is also confirmed by: (a) Figs. 21 and 22 that show (accuracy vs. iterations)and (loss vs. iterations) for CT dataset; (b) Figs. 23 and 24 that show (accuracy vs. iterations) and (loss vs. iterations) for X-ray dataset; and (c) the ROC curves shown in Figs. 25 and 26 for both CT and X-ray datasets, respectively.
Finally, Table 11 provides a comparison between the proposed DFC approach and the state-of-the-art methodologies. These results also prove the superiority of the proposed DFC approach in accurately diagnosing the COVID-19 disease.
5 Conclusions
The COVID-19 has been considered as a life-threatening disease that affects lungs. Hence, the rapid diagnosis of COVID-19 has become a vital necessity. In this paper, a CNN-based DFC approach has been presented. It adopts the concatenation process of the extracted deep features of X-ray and CT images. The proposed approach has been implemented in two different ways. In the first way, X-ray and CT image features are extracted by the proposed CNN. Then, these features are combined to form the classification descriptor. This way has achieved an accuracy of \(96.13\%\), a precision of \(94.37\%\), a recall of \(97.04\%\), and an f_score of \(95.69\%\). The second way has been mainly intended to enhance the results for either X-ray or CT dataset. This could be accomplished by using three CNNs (the proposed CNN, GoogleNet, and ResNet-18) to extract features. Then, the DFC has been applied on these features to obtain the classification descriptor. For CT images, this way has achieved an accuracy of \(98.9\%\), a precision of \(99.36\%\), a recall of \(98.5\%\), and an f_score of \(98.92\%\). For X-ray images, this way has attained an accuracy of \(99.3\%\), a precision of \(99.79\%\), a recall of \(98.8\%\), and an f_score of \(99.3\%\). Hence, the proposed approach can be used efficiently for the diagnosis of COVID-19 cases from medical images.
References
Amato F, Cozzolino G, Mazzeo A, Romano S (2018) Intelligent medical record management: a diagnosis support system. Int J High Perform Comput Netw 12(4):391–399
Braman N, Prasanna P, Whitney J, Singh S, Beig N, Etesami M, Bates DD, Gallagher K, Bloch BN, Vulchi M et al (2019) Association of peritumoral radiomics with tumor biology and pathologic response to preoperative targeted therapy for her2 (erbb2)-positive breast cancer. JAMA Netw Open 2(4):e192561–e192561
Chen XW, Lin X (2014) Big data deep learning: challenges and perspectives. IEEE Access 2:514–525
Chen T, ChefdHotel C (2014) Deep learning based automatic immune cell detection for immunohistochemistry images. In: International workshop on machine learning in medical imaging, Springer, pp 17–24
Cheng JZ, Ni D, Chou YH, Qin J, Tiu CM, Chang YC, Huang CS, Shen D, Chen CM (2016) Computer-aided diagnosis with deep learning architecture: applications to breast lesions in us images and pulmonary nodules in ct scans. Sci Rep 6(1):1–13
Dorgham O, Al-Rahamneh B, Almomani A, Khatatneh KF et al (2018) Enhancing the security of exchanging and storing dicom medical images on the cloud. Int J Cloud Appl Comput (IJCAC) 8(1):154–172
Du C, Wang Y, Wang C, Shi C, Xiao B (2020) Selective feature connection mechanism: concatenating multi-layer cnn features with a feature selector. Pattern Recogn Lett 129:108–114
El-Shafai W, El-Samie FA (2020) Going deeper with convolutions. In: Extensive and Augmented COVID-19 X-Ray and CT Chest Images Dataset. https://doi.org/10.17632/8h65ywd2jr.2
Esteva A, Kuprel B, Novoa RA, Ko J, Swetter SM, Blau HM, Thrun S (2017) Dermatologist-level classification of skin cancer with deep neural networks. Nature 542(7639):115–118
Farooq M, Hafeez A (2020) Covid-resnet: a deep learning framework for screening of covid19 from radiographs. arXiv preprint arXiv:200314395
Ghoneim A, Muhammad G, Amin SU, Gupta B (2018) Medical image forgery detection for smart healthcare. IEEE Commun Mag 56(4):33–37
Guo P, Evans A, Bhattacharya P (2018) Nuclei segmentation for quantification of brain tumors in digital pathology images. Int J Softw Sci Comput Intell (IJSSCI) 10(2):36–49
Hall LO, Paul R, Goldgof DB, Goldgof GM (2020) Finding covid-19 from chest x-rays using deep learning on a small dataset. arXiv preprint arXiv:200402060
He K, Zhang X, Ren S, Sun J (2016) Deep residual learning for image recognition. In: Proceedings of the IEEE conference on computer vision and pattern recognition, pp 770–778
Huang C, Wang Y, Li X, Ren L, Zhao J, Hu Y, Zhang L, Fan G, Xu J, Gu X et al (2020) Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. The Lancet 395(10223):497–506
Huang G, Liu Z, Van Der Maaten L, Weinberger KQ (2017) Densely connected convolutional networks. In: Proceedings of the IEEE conference on computer vision and pattern recognition, pp 4700–4708
Iandola FN, Han S, Moskewicz MW, Ashraf K, Dally WJ, Keutzer K (2016) Squeezenet: alexnet-level accuracy with 50x fewer parameters and< 0.5 mb model size. arXiv preprint arXiv:160207360
Jin S, Wang B, Xu H, Luo C, Wei L, Zhao W, Hou X, Ma W, Xu Z, Zheng Z, et al. (2020) Ai-assisted ct imaging analysis for covid-19 screening: building and deploying a medical ai system in four weeks. MedRxiv
Jmour N, Zayen S, Abdelkrim A (2018a) Convolutional neural networks for image classification. In: 2018 International Conference on Advanced Systems and Electric Technologies (IC\_ASET), IEEE, pp 397–402
Jmour N, Zayen S, Abdelkrim A (2018b) Convolutional neural networks for image classification. In: 2018 International Conference on Advanced Systems and Electric Technologies (IC\_ASET), IEEE, pp 397–402
Krizhevsky A, Sutskever I, Hinton GE (2012) Imagenet classification with deep convolutional neural networks. Adv Neural Inform Process Syst 25:1097–1105
LeCun Y, et al. (2015) Lenet-5, convolutional neural networks 20(5):14 http://yann.lecun.com/exdb/lenet
Leymarie F, Levine MD (1993) Tracking deformable objects in the plane using an active contour model. IEEE Trans Pattern Anal Mach Intell 15(6):617–634
Liang X, Hu P, Zhang L, Sun J, Yin G (2019) Mcfnet: multi-layer concatenation fusion network for medical images fusion. IEEE Sens J 19(16):7107–7119
Li L, Qin L, Xu Z, Yin Y, Wang X, Kong B, Bai J, Lu Y, Fang Z, Song Q et al (2020) Artificial intelligence distinguishes covid-19 from community acquired pneumonia on chest ct. Radiology
Ma C, Mu X, Sha D (2019) Multi-layers feature fusion of convolutional neural network for scene classification of remote sensing. IEEE Access 7:121685–121694
Michailovich O, Rathi Y, Tannenbaum A (2007) Image segmentation using active contours driven by the bhattacharyya gradient flow. IEEE Trans Image Process 16(11):2787–2801
Najafabadi MM, Villanustre F, Khoshgoftaar TM, Seliya N, Wald R, Muharemagic E (2015) Deep learning applications and challenges in big data analytics. J Big Data 2(1):1–21
Narin A, Kaya C, Pamuk Z (2020) Automatic detection of coronavirus disease (covid-19) using x-ray images and deep convolutional neural networks. arXiv preprint arXiv:200310849
Nguyen LD, Lin D, Lin Z, Cao J (2018) Deep cnns for microscopic image classification by exploiting transfer learning and feature concatenation. In: 2018 IEEE International Symposium on Circuits and Systems (ISCAS), IEEE, pp 1–5
Noreen N, Palaniappan S, Qayyum A, Ahmad I, Imran M, Shoaib M (2020) A deep learning model based on concatenation approach for the diagnosis of brain tumor. IEEE Access 8:55135–55144
Qu Z, He H, Liu W, Ma S (2019) A self-adaptive quantum steganography algorithm based on qlsb modification in watermarked quantum image. Int J High Perform Comput Network 14(2):121–129
Rahimzadeh M, Attar A (2020) A modified deep convolutional neural network for detecting covid-19 and pneumonia from chest x-ray images based on the concatenation of xception and resnet50v2. Inform Med Unlock 19:100360
Sajjad M, Khan S, Muhammad K, Wu W, Ullah A, Baik SW (2019) Multi-grade brain tumor classification using deep cnn with extensive data augmentation. J Comput Sci 30:174–182
Sandler M, Howard A, Zhu M, Zhmoginov A, Chen LC (2018) Mobilenetv2: Inverted residuals and linear bottlenecks. In: Proceedings of the IEEE conference on computer vision and pattern recognition, pp 4510–4520
Sengupta A, Ye Y, Wang R, Liu C, Roy K (2019) Going deeper in spiking neural networks: Vgg and residual architectures. Front Neurosci 13:95
Shi F, Xia L, Shan F, Wu D, Wei Y, Yuan H, Jiang H, Gao Y, Sui H, Shen D (2020) Large-scale screening of covid-19 from community acquired pneumonia using infection size-aware classification. arXiv preprint arXiv:200309860
Singhal T (2020) A review of coronavirus disease-2019 (covid-19). Indian J Pediatr 87(4):281–286
Singh K, Gupta G, Vig L, Shroff G, Agarwal P (2017) Deep convolutional neural networks for pairwise causality. arXiv preprint arXiv:170100597
Song Y, Zheng S, Li L, Zhang X, Zhang X, Huang Z, Chen J, Zhao H, Jie Y, Wang R, et al. (2020) Deep learning enables accurate diagnosis of novel coronavirus (covid-19) with ct images. MedRxiv
Szegedy C, Liu W, Jia Y, Sermanet P, Reed S, Anguelov D, Erhan D, Vanhoucke V, Rabinovich A (2015a) Going deeper with convolutions. In: Proceedings of the IEEE conference on computer vision and pattern recognition, pp 1–9
Szegedy C, Liu W, Jia Y, Sermanet P, Reed S, Anguelov D, Erhan D, Vanhoucke V, Rabinovich A (2015b) Going deeper with convolutions. In: Proceedings of the IEEE conference on computer vision and pattern recognition, pp 1–9
Tan M, Le Q (2019) Efficientnet: rethinking model scaling for convolutional neural networks. In: International Conference on Machine Learning, PMLR, pp 6105–6114
Xie X, Zhong Z, Zhao W, Zheng C, Wang F, Liu J (2020a) Chest ct for typical coronavirus disease 2019 (covid-19) pneumonia: relationship to negative rt-pcr testing. Radiology 296(2):E41–E45
Xie X, Zhong Z, Zhao W, Zheng C, Wang F, Liu J (2020b) Chest ct for typical coronavirus disease 2019 (covid-19) pneumonia: relationship to negative rt-pcr testing. Radiology 296(2):E41–E45
Xu X, Jiang X, Ma C, Du P, Li X, Lv S, Yu L, Ni Q, Chen Y, Su J et al (2020) A deep learning system to screen novel coronavirus disease 2019 pneumonia. Engineering 6(10):1122–1129
Zheng Q, Wang X, Khan MK, Zhang W, Gupta BB, Guo W (2017) A lightweight authenticated encryption scheme based on chaotic scml for railway cloud service. IEEE Access 6:711–722
Zheng C, Deng X, Fu Q, Zhou Q, Feng J, Ma H, Liu W, Wang X (2020) Deep learning-based detection for covid-19 from chest ct using weak label. MedRxiv
Zhu N, Zhang D, Wang W, Li X, Yang B, Song J, Zhao X, Huang B, Shi W, Lu R, et al. (2020) A novel coronavirus from patients with pneumonia in china, 2019. N Engl J Med
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Saad, W., Shalaby, W.A., Shokair, M. et al. COVID-19 classification using deep feature concatenation technique. J Ambient Intell Human Comput 13, 2025–2043 (2022). https://doi.org/10.1007/s12652-021-02967-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12652-021-02967-7