Abstract
This article reports on the application of organically modified silica (ORMOSIL) nanoparticles as an efficient in vitro gene delivery system in the recent years. Based on that prime objective, the present study addresses the possible ways to reduce cancers incidence at cellular level. In this context, ORMOSIL nanoparticles had been synthesized and incubated along with pCMV–Myc (3.8 kb) plasmid vector construct carrying p53gene, and transfected into the breast cancer cell line MCF-7 cells. Western blot analysis showed that the p53 protein was significantly expressed in breast cancer cell upon transfection. The confocal and electron microscopic studies further confirmed that the nanoparticles were accumulated in the cytoplasm and the nucleus of the cancer cells transfected with p53 gene. Interesting agarose gel electrophoresis studies revealed that the nanoparticles efficiently complex with pCMV–Myc vector. The anti-cancer properties of p53 were demonstrated by assessing the cell survival and growth rate which showed a positive linear correlation in cancer cells. Whereas, the growth rate was significantly reduced in ORMOSIL/p53/pCMV–Myc transfected breast cancer cells compared to the growth rate of non-transfected cells. The results of this approach using ORMOSIL nanoparticles as a non-viral gene delivery platform have a promising future for use as effective transfection agent for therapeutic manipulation of cancer cells and targeted cancer gene therapy in vivo.
Similar content being viewed by others
1 Introduction
Nanomedicine is an emerging new field created by the combination of nanotechnology and medical sciences (Roy et al. 2005). The application of nanotechnology to biomedical research is expected to have a major impact in leading the development of new types of diagnostic and therapeutic tools (Prasad 2003, 2004). One focus in nanobiotechnology is the development and use of non-viral vectors for safe and efficient gene delivery (Davis 1997; Anderson 1992, 1998). With the potential of gene transfer as a therapeutic tool, the application of nanotechnology in the development of non-viral transfection agents for gene therapy is need of the hour (Bharli et al. 2005). The chances of using viral vectors as gene carriers is limited because of the risk factors such as pathogenicity, immunogenicity etc., and there is a need to supply a sufficient amount of DNA to the surface of the cells for effective gene transfer (Luo and Saltzman 2000). In spite of these limitations, the research focus has shifted towards the development of nanoparticle dependent gene delivery system. The other major advantages of nanoparticle vectors were the absence of limitations on the size and number of gene inserts. Ultrafine silica nanoparticles, with surfaces functionalized by cationic-amino groups, have been shown to not only bind and protect plasmid DNA from enzymatic degradation but also to transfect efficiently cultured cells and express encoded proteins (Allister et al. 2002; Kneuer et al. 2000a, b). Silica nanoparticles modified with aminosilanes [either N-(2 aminoethyl)-3 aminopropyltrimethoxysilane or N-(6-aminohexyl)-3 aminopropyltrimethoxysilane] are able to condense and deliver DNA, very much like cationic polymers (He et al. 2003; Kneuer et al. 2000a, b), without the addition of other cationic transfection reagents (Kneuer et al. 2000a, b). Other cationic silica nanoparticles with surfaces modified by aminohexyl-aminopropyltrimethoxysilane (AHAPS) are also reported to be successful transfection reagents (Ravi Kumar et al. 2004). Organically modified silica (ORMOSIL) nanoparticles have a potential to overcome many limitations of their “unmodified” silica counterparts. The presence of both hydrophobic and hydrophilic groups on the precursor alkoxyorganosilane helps them to self-assemble both as normal micelles and reverse micelles under appropriate conditions. The resulting micelles (and reverse micelles) cores can be loaded with bio-molecules such as drugs, proteins, etc. (Bharali et al. 2003). Such a system has number of advantages: (1) they can be loaded with either hydrophilic or hydrophobic drugs/dyes; (2) they can be precipitated in oil-in-water microemulsions in which corrosive solvents such as cyclohexane and complex purification steps such as solvent evaporation, ultracentrifugation, etc. can be avoided; (3) their organic groups can be modified further for attachment of targeting molecules; and (4) they can be possibly biodegraded through the biochemical decomposition of the Si–C bond (Bacskai et al. 2003). The presence of the organic group also imparts some degree of flexibility to the otherwise rigid silica matrix, which is expected to enhance the stability of such particles in an aqueous system against precipitation. In the present work, the hybrid amino-functionalized ORMOSIL nanoparticles have been synthesized by a synchronous hydrolysis of vinyltriethoxysilane (VTES) and 3-aminopropyltriethoxysilane (APTES). By varying the concentrations of Aerosol-OT and VTES, monodispersed nanoparticles of the mean diameter of the ORMOSILNs were under 60 nm which are highly stable in aqueous conditions (Das et al. 2002). Furthermore, the nanoparticles have been characterized by zeta potential measurement, XRD, and FTIR. The amino-functionalized nanoparticles were able to electrostatically condense DNA (both plasmid and genomic) and protect it from enzymatic degradation. Using confocal microscopy, it was found that the fluorescently labeled nanoparticles extensively accumulated in the cytoplasm and the nucleus of cancer cell lines used for the study (Jain et al. 1998). It was found that the nanoparticles release the genetic material inside the cytoplasm, which diffuses to the nucleus, a prerequisite for successful gene therapy. The present study clearly demonstrates the transfection efficiency of ORMOSIL nanoparticles as non-viral vectors, which are shown by the expression of p53 genes in the breast carcinoma cell lines.
2 Experimental
2.1 Materials
ORMOSIL precursors are surfactant (Aerosol-OT 98 %) and co-surfactant (n-butanol 99.8 %), vinyltriethoxysilane (VTES 97 %), 3-aminopropyltriethoxysilane (APTES 99 %), and dimethyl sulfoxide (DMSO 99.5 %). The human cancer cell lines such as MCF-7 were generously donated by King Institute of Preventive Medicine Chennai (Dr. P. Gunasekaran’s laboratory). DMEM was purchased from Sigma, USA. The cells were seeded into 25-mm culture flasks and incubated at 37°C, in 5 % CO2. pCMV plasmid construct carrying anti-cancer protein p53 was purchased from Clontech. GFP was purchased from Invitrogen (Bangalore, India).
2.2 Preparation of ORMOSIL nanoparticles
The nanoparticles were synthesized in the non-polar core of Aerosol-OT/DMSO/water micelles as reported by Roy et al. and is briefly outlined as follows: Typically, the micelles were prepared by dissolving 0.44 g Aerosol-OT and 800 μl n-butanol in 20 ml of double-distilled water by vigorous magnetic stirring. One hundred microliters of DMSO was added. After that, 200 μl of neat VTES was added to the micelles system and the resulting solution was stirred for ~30 min. Furthermore, the ORMOSIL nanoparticles were precipitated by adding aqueous ammonia solution or APTES and stirring for ~20 h at room temperature using a magnetic stirrer. Formation of the nanoparticles is indicated by a white blue color. Surfactant Aerosol-OT and co-surfactant n-butanol were removed by dialyzing the aqueous solution against distilled water in a 12- to 14-kDa cut-off cellulose membrane for 50 h. The dialyzed solution was filtered through a 0.2-μm cut-off membrane filter and used straightway for further experimentation.
2.3 Characterization of ORMOSIL nanoparticles
Transmission electron microscopy was used to determine the morphology and the size of the aqueous dispersion of nanoparticles with a JEOL JEM 2020 electron microscope operating at an accelerating voltage of 200 kV. X-ray diffraction using Cukα radiation (PAN analytical X’pert Pro MPD diffractometer) was used to determine the amorphous structure of ORMOSIL nanoparticles. Powder X-ray analysis was carried out using a Philips Model PW 1050/37 diffractometer, operating at 40 kV and 30 mA, with a step size of 0.02° (2θ). Dried and powdered samples of colloids with the stabilizer before and after heat treatment were used for the measurements. The surface groups of the nanoparticles were qualitatively confirmed using FTIR spectroscopy. FTIR spectra were recorded on a Perkin-Elmer spectrum 2000 FTIR spectrophotometer. The pellets of the lyophilized powder were made with dried KBr. The size distribution of ORMOSILNs alone and their complexes with various amounts of DNA was measured by quasi-elastic light scattering (QLS) using Nicomp-370 (Nicomp Particles Sizing System, Santa Barbara, USA). For particle size analysis of the complexes, the complexes were diluted with phosphate-buffered saline (PBS) just prior to size measurement. The change in the particle diameter of complexes was measured for 3 min using Gaussian analysis. The zeta potential of complexes was analyzed using an electrophoretic light scattering spectrophotometer (ELS-8000, OTSUKA Electronics Co. Ltd., Japan) at room temperature to monitor the electrophoretic mobility of transfected complexes.
2.4 Nano-DNA complex preparation
We examined the complex formation of plasmid DNA with nanoparticles by agarose gel electrophoresis. DNA loading of ORMOSIL nanoparticles was accomplished by incubation with pCMV/p53 for 30 min at room temperature. The resulting ORMOSIL/pCMV/p53 nanoparticles were suspended in phosphate-buffered saline (PBS; 140 μg/ml DNA). The complex formation were run on 1 % agarose at 100 V for 1.5 h, subsequently stained with ethidium bromide, and documented by using a gel documentation system. Genei Ultraviolet Benchtop transilluminator was used in conjugation with an Olympus Digital Camedia C-4000 Zoom color camera with a UV filter and lens. The documentation was completed by using the DOC-IT system software.
2.5 In vitro transfection
MCF-7 (human breast carcinoma) cells were seeded in 24-well tissue culture plates at a density of 2 × 105 cells/well. The cells were allowed to adhere overnight as a monolayer and to achieve 70–80 % confluence. Transfection complexes were prepared by mixing the plasmid DNA (p53–pCMV) with ORMOSILNs and were incubated for 15 min at room temperature before in vitro studies. Prior to transfection, 800 μl of serum-free DMEM was added to each well; 100 μl of transfection complexes mixed with 2 μg of the plasmid DNA and various amounts (6–48 μg) of ORMOSILNs were then added to each of the wells. After 5 h of incubation at 37°C under 5 % CO2, cells were rinsed and cultured for 24 h in 1 ml of medium containing 10 % (FBS).
2.6 Western blot analysis
The levels of p53 protein expression in transfected cells with the plasmid DNA/ORMOSILNs complexes were determined by Western blot analysis. The transfected cells were harvested and lysed in lysis buffer (150 mM NaCl; 20 mM Tris base, pH 7.5; 1 mM PMSF; 1 mM Na3VO4; 25 mM NaF; 1 % aprotinin; 10 μg/ml leuprotinin; 1 % Triton X-100; 1 % NP-40) on ice for 30 min. After samples were centrifuged at 12,000 rpm for 10 min, the protein concentrations of supernatants were determined by Bio-Rad DC (detergent-compatible) microprotein assay using bovine serum albumin (BSA) as a protein standard. Then 15-μg aliquots of proteins were separated by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS–PAGE) and transferred to nitrocellulose filters. The nitrocellulose membrane was incubated for 1 h in a blocking buffer (5 % non-fat milk in PBS) followed by incubation with the mouse anti-human p53 antibody (Pab240, Santa Cruz, CA, USA).
2.7 Cell viability
The effects of the plasmid DNA/ORMOSILNs complexes on growth inhibition were determined using a cell growth assay. The MCF-7 cells were seeded in six-well plates at a density of 1 × 104 cells/well. After 24 h, the cells were transfected with the plasmid DNA alone, DNA/ORMOSILNs, or DNA/Lipofectin® complexes, respectively. After transfection, the amounts of viable adherent cells were determined by trypan blue exclusion assay performed every other day. Untreated cells were used as a control. Growth assays were performed in triplicate.
2.8 Statistical analysis
Statistical analysis of data was performed using Student’s t test and analysis of variance (ANOVA). A p value less than 0.05 were considered significant.
3 Results and discussion
3.1 Physicochemical characteristics of ORMOSILNs
Figure 1 shows transmission electron microscopy images of ORMOSIL nanoparticles. Note that the particles are spherical in shape and highly monodispersed. For the preparation of cationic ORMOSILNs, VTES was selected as a silica component that would constitute the core of the ORMOSILNs. Aerosol-OT, DMSO, and APTES were used as cationic ORMOSIL, helper VTES, and surfactant, respectively. The mean diameter of the ORMOSILNs was under 60 nm, which is generally known to be an effective size for high transfection efficiency. The zeta potentials of ORMOSILNs and ORMOSILNs/DNA were approximately 8 and 13 mV, respectively. To investigate whether the changes in the packing material or storage conditions may further improve the stability of freeze-dried ORMOSILNs, ORMOSILNs were freeze-dried using sucrose as a cryoprotectant, and the particle diameter and transfection efficiency were subsequently determined. The particle diameters of ORMOSILNs and ORMOSILNs/DNA were slightly increased but both of them were under 60 nm. We also compared the transfection efficiencies of the ORMOSILNs before freeze drying (Table 1).
3.2 X-ray diffraction analysis
The vinyltriethoxysilane (VTES) 3-aminopropyltriethoxysilane (APTES) capped ORMOSIL nanoparticles (Fig. 2) were investigated by X-ray diffraction technique. In the selected constituent concentration, spherical ORMOSIL nanoparticle can be obtained as colloidal ORMOSIL nanoparticles that are based on the hydrolysis and polycondensation of VTES in the aqueous core of the reverse micelles droplets. The reaction continued until the solution is supersaturated. To investigate the possibility of tailoring the particle size distribution, a kinetic study of the particle size evolution as a function of reaction time was carried out. The ORMOSILNs were amorphous according to XRD with peaks on 2θ = 220 conforming to the JCPDS parameters 27.1402. This demonstrated that a high percentage of these particles are amorphous besides a few of them that were crystalline in nature (Bolton and Kearns 1978; Vacassy et al. 2000).
3.3 FTIR analysis
The nanoparticle surface design involves an optimum balance of the use of inert and active surface functional groups to achieve minimal nanoparticle aggregation and reduce nanoparticle non-specific binding. Silica nanoparticles were prepared in a water-in-oil microemulsion and subsequently surface-modified via co-hydrolysis with tetraethylorthosilicate (TEOS) and organosilane reagents. Nanoparticles with different functional groups, including amine and amine/phosphonate, were produced in the present investigation. The FT-IR spectrum (Fig. 3) of synthesized hybrid amino-functionalized ORMOSIL nanoparticles by a synchronous hydrolysis of VTES and 3-aminopropyltriethoxysilane (APTES) sample shows peaks around 1,730 and 3,516 cm−1 corresponding to the carboxyl and hydroxyl groups (Shan et al. 2000). The IR spectrum shows the peaks for NH2 group as well as silica. The amino-functionalized nanoparticles are able to electrostatically condense DNA (both gene of interest and genomic) and protect it from enzymatic degradation. Hence, silica was functionalized with amino group in the present investigation. Furthermore, in the given IR spectrum the amino group peaks were noticed between 3,000 and 3,600 (stretched vibration) and also between 1,550 and 1,640 (bending vibration). Interestingly, the peak for silica is observed between 1,000 and 1,250.
Respectively, the FT-IR peaks belong to the Si–O stretching vibration of Si–OH bond. A strong peak near 1,046, 773, and 467 cm−1 has been assigned to Si–O Si stretching bond which implies the condensation of silicon alkoxide in monomer vinyltriethoxysilane (VTES). The presence of the vinyl group in VTES can usually be recognized by the presence of a minimum intensity of broad band at 3,516 cm−1 due to the unsaturated =C–H stretching vibration. In plane bending vibration of the group =C −H−H in the presence of VTES peak at 1,406 cm−1 and C=C stretching vibration give shift to an absorption at 1,646 cm−1 (Vogel 1989). The two bands at 2,959 and 2,870 cm−1 were assigned to the asymmetric and symmetric stretching vibration modes of the CH3 and CH2 groups in presence of VTES and APTES (Sharma et al. 2004). Rocking vibration of the CH3 groups at 1,165 cm−1 was present in VTES, and Si–C bands at 773 and 1,218 cm−1 peaks were seen (Table 2).
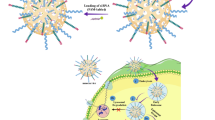
3.4 Examination of the complex formation of plasmid DNA with nanoparticles
For non-viral gene delivery, we have developed and tested the formation of nanoparticles which consist of a stable aqueous dispersion of ultrafine ORMOSIL nanoparticles. ORMOSIL-based nanoparticles can be synthesized with varying surface change and are stable (non-aggregating) in aqueous environments. The genetic payload (negative charged DNA) is bound to the cationic group present on the surface of the nanoparticles. The non-aggregating potential, coupled with the designed ability to bind and protect DNA, provides a second platform from which a non-viral gene transfer vector will be produced and tested. The nanoparticles for DNA delivery are functionalized with amino group, which made it possible to electrostatically attach DNA molecules on the surface as shown schematically in Fig. 4. Nanoparticles were found to have high efficiency for the formation of ORMOSIL DNA complex. Figure 5 represents the results of agarose gel electrophoresis of plasmid DNA, free and complexes with ORMOSIL nanoparticles. It can be seen that with increasing amounts of amino groups (positive charge) on the nanoparticles, the mobility of the DNA complexes toward 50 μl, 30 μl, and 10 μl retards the positive terminal (lanes 3–5). This result suggests that the plasmid is no more able to move freely because the resulting nanoparticle–DNA complex has restricted the mobility in the gel. After treatment with DNase 1, the free plasmid is completely digested (Fig. 5, lane 6). The reason for this protection against enzymatic digestion is not yet fully understood. It was suggested recently that this behavior can be due to (1) repulsion of Mg2+ ions (which are necessary for the enzymatic reaction) by the amino groups, (2) a hindered access of the enzymes to the DNA that is immobilized on the nanoparticle surface, or (3) both (He et al. 2003). Note that the plasmid treated with the non-amino terminated particle (ORMOSIL) is also partially protected (Fig. 5, lane 2) because it has bands corresponding to both its non-enzymatically treated counterpart (Fig. 5, lane 3) and also some DNA fragments appearing at the bottom of the band. Therefore, these nanoparticles can also be considered as some kind of inhibitors toward the enzymatic action of DNase 1 on plasmid DNA. Alternatively, it is also possible that the interaction between the genetic material and the nanoparticle will not be entirely of electrostatic nature.
3.5 Western blot analysis for p53 expression
In order to evaluate the uptake of p53 genes into MCF-7 by nanoparticles, the transfected MCF-7, after 3 days of culturing, were detected via Western blotting with protein-specific antibodies (Fig. 6). As shown in the figure, protein levels of the p53 of MCF-7 were significantly increased by the ORMOSIL nanoparticles. In contrast, the protein level of the p53 genes in MCF-7 using ORMOSIL carriers was found to be highly increased. However, the expression of p53 genes in MCF-7 was slightly detected in the Lipofectin® via Western blot analysis. This means that the ORMOSIL nanoparticles predominantly expressed the p53 genes in MCF-7 as compared to the other gene carriers. Via the expression of p53 genes in MCF-7, ORMOSIL nanoparticles appear to be superior gene delivery vehicles.
3.6 In vitro growth assay
Using in vitro growth assay, the effect of the transfection of p53–pCMV/ORMOSILNs on the rate of MCF-7 cell growth was determined by measuring the doubling time compared to that of uninfected control cells. On day 0, the cell lines were passed into monolayer cultures in 24-well flat-bottomed microplates. The following day, the cells were transfected with plasmid DNA alone, DNA/ORMOSILNs, or DNA/Lipofectin®. The growth rate of p53–pCMV/ORMOSILNs-transfected cells was significantly inhibited compared with control cells or cells transfected with p53 DNA alone (Fig. 7). These results indicate that ORMSILNs-mediated p53 DNA delivery inhibited the growth of MCF-7 cells. Transfection of MCF-7 cells with p53 DNA alone had a slight effect on cell growth compared to the uninfected cells. Thus, the inhibitory p53–pCMV/ORMOSILNs transfection in MCF-7 cells is likely due to the expression of p53 protein.
The in vitro growth inhibition of the MCF-7 cells after transfection with DNA/ORMOSILNs complexes. The number of adherent cells was determined at each time point. Each point indicates the mean ± SD of values obtained from the assay of cells from six replicate microtiter plate wells. *p < 0.05, compared with the in vitro growth inhibition of the control
Efficient inhibition of cell growth by p53–pCMV/ORMOSILNs complexes indicates that these complexes would be efficient not only in increasing the expression of p53 proteins but also in inhibiting the growth of breast cancer cells.
3.7 Gene transfer efficiency of ORMOSIL nanoparticles
The efficiency of ORMOSIL nanoparticles mediated transformation was analyzed using a transmission electron microscope (TEM) at cellular level. Figure 8 shows that the ORMOSIL nanoparticles are highly dispersed in the cytoplasm of transformed breast cancer cell line MCF-7 cells. There was no cellular distortion present on TEM images of breast carcinoma cells exposed to ORMOSIL nanoparticles alone. All organelles are intact and the cells imaged are unchanged except for the presence of ORMOSIL nanoparticles. Normally for breast carcinoma cells, the nuclear cytoplasmic ratio is very low. Lower concentration of nuclear cytoplasm ratio in transformed breast carcinoma cells was also confirmed by TEM image. Furthermore, at ×100,000 magnification, ORMOSIL nanoparticles of various sizes that are dispersed in cytoplasm are clearly visible. A previous study reported that hydrophobic dye-doped ORMOSIL nanoparticles are actively taken up by tumor cells in vitro, as seen by fluorescence staining of the cytoplasm (Roy et al. 2003). This cytoplasmic staining is observed for several types of tumor cells that we have investigated, an example of which is shown for MCF-7 cells in (Fig. 9). Although the mechanism of this cellular uptake is not yet fully understood, the net positive charge on the nanoparticles has been thought to play a crucial role. A similar observation was found in a recent study that has demonstrated high non-specific uptake of anionic magnetic nanoparticles by cells in vitro (Wilhem et al. 2003). In gene delivery, the genetic material, as soon as it is released inside the cytoplasm of a cell, should migrate to the nucleus (Davis 1997). Again, optical imaging can play a significant role in studying this nuclear migration of DNA. In our experiment, we irreversibly labeled DNA with the fluorescent dye GFP and DAPI complexed with unlabeled ORMOSIL nanoparticles as described. The post-release nuclear trafficking of DNA is demonstrated in the confocal image of MCF-7 cells in Fig. 9a. After comparison of this image with the image of MCF-7 cells stained with DAPI encapsulated nanoparticles (Fig. 9b), it can be seen that in addition to the cytoplasmic staining, there is considerable staining inside the nucleus. Optimal transfection conditions were evaluated by varying the amounts of plasmid DNA at different ORMOSIL/DNA ratios, incubation times, and media in order to form the complexes. Figure 9c and d shows finally the expression of nano-DNA complex as confirmed by DAPI fluorescent stain with the help of fluorescent microscopy. The 4-h incubation of the nano-DNA complex with breast cancer cell line MCF-7 showed that the nano-DNA complex was taken up completely by the cell as compared to 2 h of incubation. We observed that pCMV–p53 was transfected successfully by using the ORMOSIL nanoparticles as the non-viral delivery vector.
Transmission electron microscopic analysis of ORMOSIL nanoparticles transfection at cellular level. a The ORMOSILNs are mainly distributed in the cytoplasm of MCF-7. The arrow indicates the ORMOSILNs. b Non-transfected cells (control). c ORMOSILNs distribute in the cytoplasm at ×100,000 magnification. The arrow indicates the ORMOSILNs in different sizes. d Non-transfected cells at ×100,000 magnification (control)
Confocal and fluorescence images of cancer cells treated with ORMOSILNs previously incubated with DNA labeled with ethidium monoazide (EMA). a Confocal bioimaging of cancer cells demonstrating expression of green fluorescent protein following in vitro administration. b Fluorescent image of nano-DNA complex stained with 4′-6-diamidino-2-phenylindole (DAPI). c 2-h incubation of nano-DNA complex with cells shows that the nanoparticles are present outside the cells. d Transformed into cells after 4-h incubation
4 Conclusions
The present study shows that the nanomedicine approach using ORMOSIL nanoparticles provides a promising direction for non-viral gene delivery. In the present context, the use of DAPI, together with fluorescence imaging, established DNA delivery to the cell nucleus, whereas the use of pCMV/p53 western blot provided evidence for the functionality of delivered DNA. Our findings provide additional information regarding the delivery of p53 gene into a selected breast cancer cell line which is perfectly expressed in the cells. Thus, the ORMOSILNs are recommended. In particular, the effective indicated that the ORMOSILNs-mediated delivery of p53 gene might show potential for clinical use in non-viral vector mediated breast cancer therapies.
References
Allister MK, Sazani P, Adam M, Cho MJ, Rubinstein M, Samulski RJ, Desimone JM (2002) Polymeric nanogels produced via inverse microemulsion polymerization as potential gene and antisense delivery agents. J Am Chem Soc 124:15198–15207
Anderson WF (1992) Human gene therapy. Science 256:808–813
Anderson WF (1998) Human gene therapy. Nature 392:25–30
Bacskai BJ, Skoch J, Hickey GA, Allen R, Hyman BT (2003) Fluorescence resonance energy transfer determinations using multiphoton fluorescence lifetime imaging microscopy to characterize amyloid-beta plaques. J Biomed Opt 8:368–375
Bharali DJ, Sahoo SK, Mozumdar S, Maitra A (2003) Cross-linked polyvinylpyrrolidone nanoparticles: a potential carrier for hydrophilic drugs. J Colloid Interface Sci 15:415–423
Bharli DJ, Klejbor I, Stachowiak EK, Dutta P, Roy I, Bergey KN, Prasad N, Stachowiak MK (2005) Organically modified silica nanoparticles: a nonviral vector for in vivo gene delivery and expression in the brain. Proc Natl Acad Sci USA 102(2005):11539–11544
Bolton PH, Kearns DR (1978) Spectroscopic properties of ethidium monoazide: a fluorescent photoaffinity label for nucleic acids. Nucleic Acids Res 5:4891–4903
Das S, Jain TK, Maitra A (2002) Inorganic–organic hybrid nanoparticles from n-octyl triethoxy silane. J Colloid Interface Sci 252:82–88
Davis SS (1997) Biomedical applications of nanotechnology—implications for drug targeting and gene therapy. Trends Biotechnol 15:217–224
He XX, Wang K, Tan W, Liu B, Lin X, He C, Li D, Huang S, Li J (2003) Bioconjugated nanoparticles for DNA protection from cleavage. J Am Chem Soc 125:7168–7169
Jain TK, Roy I, De TK, Maitra AN (1998) Nanometer silica particles encapsulating active compounds: a novel ceramic drug carrier. J Am Chem Soc 120:11092–11095
Kneuer C, Sameti M, Bakowsky U, Sciestel T, Schirra H (2000a) A nonviral DNA delivery system based on surface modified silica nanoparticles can efficiently transfect cells in vivo. Bioconjug Chem 11:926–932
Kneuer C, Sameti M, Haltner EG, Schiestel T, Schirra H, Schmidt H, Lehr CM (2000b) Silica nanoparticles modified with aminosilanes as carriers for plasmid DNA. Int J Pharm 196:257–261
Luo D, Saltzman WM (2000) Enhancement of transfection by physical concentration of DNA at the cell surface. Nat Biotechnol 18:893–895
Prasad PN (2003) Introduction to biophotonics. Wiley, New York
Prasad PN (2004) Nanophotonics. Wiley, New York
Ravi Kumar MN, Sameti M, Mohapatra SS, Kong X, Lockey RF, Bakowsky U, Lindenblatt G, Schmidt H, Lehr CM (2004) Cationic silica nanoparticles as gene carriers: synthesis, characterization and transfection efficiency in vitro and in vivo. J Nanosci Nanotechnol 4:876–881
Roy I, Ohulchanskyy TY, Pudavar HE, Bergey EJ, Oseroff AR, Morgan J, Dougherty TJ, Prasad PN (2003) Ceramic-based nanoparticles entrapping water-insoluble photosensitizing anticancer drugs: a novel drug-carrier system for photodynamic therapy. J Am Chem Soc 125:7860–7865
Roy I, Ohulchanskyy TY, Mistretta RA, Bharali DJ, Kaur N, Pudavar H, Prasad PN (2005) Optical tracking of organically modified silica nanoparticles as DNA carriers: a nonviral, nanomedicine approach for gene delivery. Proc Natl Acad Sci U S A 102:279–284
Shan Y, Gao L, Zheng S (2000) A facile approach to load CdSe nanocrystallites into mesoporous SBA-15. Mater Chem Phys 88:192–196
Sharma RK, Das S, Maitra A (2004) Surface modified ormosil nanoparticles. J Colloid Interface Sci 277:342–346
Vacassy R, Flatt RJ, Hofmann H, Choi KS, Singh RK (2000) Synthesis of microporous silica spheres. J Colloid Interface Sci 2:302–315
Vogel AL (1989) Textbook of practical organic chemistry. ELBS, UK
Wilhem C, Billotey C, Roger J, Pons JN, Bacri JC, Gazeau F (2003) Intracellular uptake of anionic superparamagnetic nanoparticles as a function of their surface coating. Biomaterials 24:1001–1011
Acknowledgments
We thank R. Paulmurugan, Department of Radiology–Diagnostic Radiology, Stanford University, for useful discussion and All India Institute of Medical Sciences New Delhi for their excellent technical support for taking confocal, electron, and fluorescent microscopy results. Other technical support was provided by Centre for Life Science DRDO Bharathiar University Coimbatore. This work is financially supported by the DST-Nanomission Division, Ministry of Science and Technology, Government of India (Ref. DST/SR/NM/NS-60/2010).
Competing interests
The authors declare that they have no competing interests
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 2.0 International License ( https://creativecommons.org/licenses/by/2.0 ), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
About this article
Cite this article
Rejeeth, C., Kannan, S. & Muthuchelian, K. Development of in vitro gene delivery system using ORMOSIL nanoparticle: Analysis of p53 gene expression in cultured breast cancer cell (MCF-7). Cancer Nano 3, 55–63 (2012). https://doi.org/10.1007/s12645-012-0030-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12645-012-0030-3