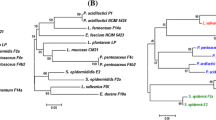
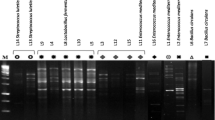
Abstract
Probiotics are live microorganisms which confer health benefits to the host. Lactic acid bacteria (LAB) are used as probiotics since decades. Enterococci being the member of LAB have proven probiotic strains; therefore, this study was aimed at finding out the potential probiotic candidates from the pool of locally isolated strains. For initial screening, one hundred and twenty-two strains were selected and subjected to different confirmatory and phenotypic tests to choose the best strains that have potential probiotic criteria, i.e., no potential virulence traits, antibiotic resistance, and having tolerance properties. Keeping this criterion, only eleven strains (n = 11) were selected for further assessment. All virulence traits such as production of hemolysin, gelatinase, biofilm, and DNase were performed and not found in the tested strains. The molecular assessment indicates the presence of few virulence-associated genes in Enterococcus faecalis strains with variable frequency. The phenotypic and genotypic assessments of antibiotic resistance profile indicate that the selected strain was susceptible to ten commonly used antibiotics, and there were no transferrable antibiotic resistance genes. The presence of CRISPR-Cas genes also confirmed the absence of antibiotic resistance genes. Various enterocin-producing genes like EntP, EntB, EntA, and EntQ were also identified in the selected strains which make them promising probiotic lead strains. Different tolerance assays like acid, NaCl, and gastric juice tolerance that mimic host conditions was also evaluated by providing artificial conditions. Cellular adhesion and aggregation properties like auto- and co-aggregation were also checked and their results reflect all in the favor of lead probiotic strains.






Similar content being viewed by others
Data Availability
All data generated or analyzed during this study are included in this published article and the supplementary information available on-line at the publisher website.
References
García-Solache M, Rice LB (2019) The Enterococcus: a model of adaptability to its environment. Clin Microbiol Rev 32(2):00058–00118. https://doi.org/10.1128/CMR.0005818
Fisher K, Phillips C (2009) The ecology, epidemiology and virulence of Enterococcus. Microbiology 155(6):1749–1757. https://doi.org/10.1099/mic.0.026385-0
Rehman M, Hasan KA, Bin-Asif H, Akram S, Zahid S, Mirani ZA, Ali SA (2021) Differentiating Enterococcus lineages in combined sewer overflow and potable water combating to hospital acquired high-level β-lactam resistance. Environ Chall 4:100094. https://doi.org/10.1016/j.envc.2021.100094
Hwanhlem N, Ivanova T, Biscola V, Choiset Y, Haertlé T (2017) Bacteriocin producing Enterococcus faecalis isolated from chicken gastrointestinal tract originating from Phitsanulok, Thailand: isolation, screening, safety evaluation and probiotic properties. Food Control 78:187–195. https://doi.org/10.1016/j.foodcont.2017.02.060
Ali SA, Bin-Asif H, Hasan KA, Rehman M, Abbasi A (2017) Molecular assessment of virulence determinants, hospital associated marker (IS16gene) and prevalence of antibiotic resistance in soil borne Enterococcus species. Microb Pathog 1(105):298–306. https://doi.org/10.1016/j.micpath.2017.02.041
Fugaban JII, Holzapfel WH, Todorov SD (2021) Probiotic potential and safety assessment of bacteriocinogenic Enterococcus faecium strains with antibacterial activity against Listeria and vancomycin-resistant enterococci. Curr Res Microb Sci 2:100070. https://doi.org/10.1016/j.crmicr.2021.100070
Moreno MF, Sarantinopoulos P, Tsakalidou E, De Vuyst L (2006) The role and application of enterococci in food and health. Int J Food Microbiol 106(1):1–24. https://doi.org/10.1016/j.ijfoodmicro.2005.06.026
Tollu G, Ekin İ (2020) Biotyping and antimicrobial susceptibility of Enterococcus faecalis and E. faecium isolated from urine and stool samples. Jundishapur J Microbiol 13(10):1–8. https://doi.org/10.5812/jjm.105136
Ben Braiek O, Smaoui S (2019) Enterococci: between emerging pathogens and potential probiotics. Biomed Res Int 2019:5938210. https://doi.org/10.1155/2019/5938210
Hasan KA, Ali SA, Rehman M, Bin-Asif H, Zahid S (2018) The unravelled Enterococcus faecalis zoonotic superbugs: emerging multiple resistant and virulent lineages isolated from poultry environment. Zoonoses Public Health 65(8):921–935. https://doi.org/10.1111/zph.12512
Bin-Asif H, Ali SA (2019) The genus Enterococcus and its associated virulent factors. Microorganisms. https://doi.org/10.5772/intechopen.89083
Plaza-Diaz J, Ruiz-Ojeda FJ, Gil-Campos M, Gil A (2019) Mechanisms of action of probiotics. Adv Nutr. https://doi.org/10.1093/advances/nmy063
Mao Q, Sun X, Sun J, Zhang F, Lv A, Hu X, Guo Y (2020) A candidate probiotic strain of Enterococcus faecium from the intestine of the crucian carp Carassius auratus. AMB Express 10(1):1–9. https://doi.org/10.1186/s13568-020-00973-0
O’Hara AM, Shanahan F (2007) Mechanisms of action of probiotics in intestinal diseases. Sci World J 7:31–46. https://doi.org/10.1100/tsw.2007.26
Nueno-Palop C, Narbad A (2011) Probiotic assessment of Enterococcus faecalis CP58 isolated from human gut. Int J Food Microbiol 145(2–3):390–394. https://doi.org/10.1016/j.ijfoodmicro.2010.12.029
Singh K, Kallali B, Kumar A, Thaker V (2011) Probiotics: a review. Asian Pac J Trop Biomed 1(2):S287–S290. https://doi.org/10.1016/S2221-1691(11)60174-3
Ohland CL, MacNaughton WK (2010) Probiotic bacteria and intestinal epithelial barrier function. Am J Physiol - Gastrointest Liver Physiol 298(6):G807–G819. https://doi.org/10.1152/ajpgi.00243.2009
Kerry RG, Patra JK, Gouda S, Park Y, Shin HS, Das G (2018) Benefaction of probiotics for human health: a review. J Food Drug Anal 26(3):927–939. https://doi.org/10.1016/j.jfda.2018.01.002
Das DJ, Shankar A, Johnson JB, Thomas S (2020) Critical insights into antibiotic resistance transferability in probiotic Lactobacillus. Nutrition 69:110567. https://doi.org/10.1016/j.nut.2019.110567
Anadón A, Martínez-Larrañaga MR, Martínez MA (2006) Probiotics for animal nutrition in the European Union. Regulation and safety assessment. Regul Toxicol Pharmacol 45(1):91–95. https://doi.org/10.1016/j.yrtph.2006.02.004
de Melo Pereira GV, de Oliveira Coelho B, Júnior AIM, Thomaz-Soccol V, Soccol CR (2018) How to select a probiotic? Biotechnol Adv 36(8):2060–2076. https://doi.org/10.1016/j.biotechadv.2018.09.003
Kechagia M, Basoulis D, Konstantopoulou S, Dimitriadi D, Gyftopoulou K, Skarmoutsou N, Fakiri EM (2013) Health benefits of probiotics: a review. ISRN Nutr 2013:1–7. https://doi.org/10.5402/2013/481651
Franz CM, Huch M, Abriouel H, Holzapfel W, Gálvez A (2011) Enterococci as probiotics and their implications in food safety. Int J Food Microbiol 151(2):125–140. https://doi.org/10.1016/j.ijfoodmicro.2011.08.014
Ferchichi M, Sebei K, Boukerb AM, Karray-Bouraoui N, Chevalier S, Feuilloley MG, Zommiti M (2021) Enterococcus spp.: is it a bad choice for a good use—a conundrum to solve. Microorganisms 9(11):2222. https://doi.org/10.3390/microorganisms9112222
Hanchi H, Mottawea W, Sebei K, Hammami R (2018) The genus Enterococcus: between probiotic potential and safety concerns—an update. Front Microbiol 9:1–16. https://doi.org/10.3389/fmicb.2018.01791
Suvorov A, Ermolenko E, Alechina G, Chernysh A, Karaseva A, Di Pierro F et al (2019) Enterococcus as probiotics: what is the advantage? Nutrafoods 1:17–25. https://doi.org/10.17470/NF-019-0003
Ali SA, Hasan KA, Bin Asif H, Abbasi A (2014) Environmental enterococci: I. Prevalence of virulence, antibiotic resistance and species distribution in poultry and its related environment in Karachi, Pakistan. Lett Appl Microbiol 58(5):423–432. https://doi.org/10.1111/lam.12208
Valledor SJD, Dioso CM, Bucheli JEV, Park YJ, Suh DH, Jung ES, Todorov SD (2022) Characterization and safety evaluation of two beneficial, enterocin-producing Enterococcus faecium strains isolated from kimchi, a Korean fermented cabbage. Food Microbiol 102:103886. https://doi.org/10.1016/j.fm.2021.103886
Andreou LV (2013) Preparation of genomic DNA from bacteria. Methods Enzymol 529:143–151. https://doi.org/10.1016/B978-0-12-418687-3.00011-2
Zahid S, Bin-Asif H, Hasan KA, Rehman M, Ali SA (2017) Prevalence and genetic profiling of tetracycline resistance (Tet-R) genes and transposable element (Tn916) in environmental Enterococcus species. Microb Pathog 111:252–261. https://doi.org/10.1016/j.micpath.2017.09.009
Uymaz Tezel B (2019) Preliminary in vitro evaluation of the probiotic potential of the bacteriocinogenic strain Enterococcus lactis PMD74 isolated from ezine cheese. J Food Qual 2019:2019. https://doi.org/10.1155/2019/4693513
Ahmadova A, Todorov SD, Choiset Y, Rabesona H, Zadi TM, Kuliyev A, Haertlé T (2013) Evaluation of antimicrobial activity, probiotic properties and safety of wild strain Enterococcus faecium AQ71 isolated from Azerbaijani Motal cheese. Food Control 30(2):631–641. https://doi.org/10.1016/j.foodcont.2012.08.009
Nami Y, Vaseghi Bakhshayesh R, Mohammadzadeh Jalaly H, Lotfi H, Eslami S, Hejazi MA (2019) Probiotic properties of Enterococcus isolated from artisanal dairy products. Front Microbiol 10:300. https://doi.org/10.3389/fmicb.2019.00300
Rehaiem A, Belgacem ZB, Edalatian MR, Martínez B, Rodríguez A, Manai M, Guerra NP (2014) Assessment of potential probiotic properties and multiple bacteriocin encoding-genes of the technological performing strain Enterococcus faecium MMRA. Food Control 37(1):343–350. https://doi.org/10.1016/j.foodcont.2013.09.044
Lindenstrauß AG, Pavlovic M, Bringmann A, Behr J, Ehrmann MA, Vogel RF (2011) Comparison of genotypic and phenotypic cluster analyses of virulence determinants and possible role of CRISPR elements towards their incidence in Enterococcus faecalis and Enterococcus faecium. Syst Appl Microbiol 34(8):553–560. https://doi.org/10.1016/j.syapm.2011.05.002
Clinical and Laboratory Standards Institute (CLSI) (2018) Performance Standards for Antimicrobial Susceptibility Testing. CLSI Approved Standard M100-S15. Clinical and Laboratory Standards Institute, Wayne
Berreta A, Baumgardner RM, Kopper JJ (2020) Evaluation of commercial veterinary probiotics containing enterococci for transferrable vancomycin resistance genes. BMC Res Notes 13(1):1–6. https://doi.org/10.1186/s13104-020-05114-1
Dziuba B, Babuchowski A, Nałęcz D, Niklewicz M (2007) Identification of lactic acid bacteria using FTIR spectroscopy and cluster analysis. Int Dairy J 17(3):183–189. https://doi.org/10.1016/j.idairyj.2006.02.013
Zahid S (2019) Molecular assessment of tetracycline resistance in environmental strains of enterococci in Karachi Pakistan. Dissertation, University of Karachi
Fu X, Lyu L, Wang Y, Zhang Y, Guo X, Chen Q, Liu C (2022) Safety assessment and probiotic characteristics of Enterococcus lactis JDM1. Microb Pathog 163:105380. https://doi.org/10.1016/j.micpath.2021.105380
Pieniz S, Andreazza R, Anghinoni T, Camargo F, Brandelli A (2014) Probiotic potential, antimicrobial and antioxidant activities of Enterococcus durans strain LAB18s. Food Control 37(1):251–256. https://doi.org/10.1016/j.foodcont.2013.09.055
Nascimento LCS, Casarotti SN, Todorov SD, Penna ALB (2019) Probiotic potential and safety of enterococci strains. Ann Microbiol 69(3):241–252. https://doi.org/10.1007/s13213-018-1412-5
Bhardwaj A, Kaur G, Gupta H, Vij S, Malik RK (2011) Interspecies diversity, safety and probiotic potential of bacteriocinogenic Enterococcus faecium isolated from dairy food and human faeces. World J Microbiol Biotechnol 27(3):59. https://doi.org/10.1007/s11274-010-0494-4
Dowdell P, Chankhamhaengdecha S, Panbangred W, Janvilisri T, Aroonnual A (2020) Probiotic activity of Enterococcus faecium and Lactococcus lactis isolated from Thai fermented sausages and their protective effect against Clostridium difficile. Probiotics Antimicrob 12:641–648. https://doi.org/10.1007/s12602-019-09536-7
Chauvière G, Coconnier MH, Kerneis S, Darfeuille-Michaud A, Joly B, Servin AL (1992) Competitive exclusion of diarrheagenic Escherichia coli (ETEC) from human enterocyte-like Caco-2 cells by heat-killed Lactobacillus. FEMS Microbiol Lett 91(3):213–217. https://doi.org/10.1111/j.1574-6968.1992.tb05211.x
Yu C, Irudayaraj J (2005) Spectroscopic characterization of microorganisms by Fourier transform infrared microspectroscopy. Biopolymers 77(6):368–377. https://doi.org/10.1002/bip.20247
Tilwani YM, Lakra AK, Domdi L, Jha N, Arul V (2022) Characterization of potential probiotic bacteria Enterococcus faecium MC-5 isolated from the gut content of Cyprinus carpio specularis. Microb Pathog 172:105783. https://doi.org/10.1016/j.micpath.2022.105783
Oruc O, Ceti̇n O, Darilmaz DO, Yüsekdag ZN (2021) Determination of the biosafety of potential probiotic Enterococcus faecalis and Enterococcus faecium strains isolated from traditional white cheeses. LWT 148:111741. https://doi.org/10.1016/j.lwt.2021.111741
Baccouri O, Boukerb AM, Farhat LB, Zébré A, Zimmermann K, Domann E, Connil N (2019) Probiotic potential and safety evaluation of Enterococcus faecalis OB14 and OB15, isolated from traditional Tunisian Testouri cheese and Rigouta, using physiological and genomic analysis. Front Microbiol 10:1–15. https://doi.org/10.3389/fmicb.2019.00881
Eaton TJ, Gasson MJ (2001) Molecular screening of Enterococcus virulence determinants and potential for genetic exchange between food and medical isolates. Appl Environ Microbiol 67(4):1628–1635. https://doi.org/10.1128/AEM.67.4.1628-1635.2001
Barbosa J, Gibbs PA, Teixeira P (2010) Virulence factors among enterococci isolated from traditional fermented meat products produced in the North of Portugal. Food Control 21(5):651–656. https://doi.org/10.1016/j.foodcont.2009.10.002
Bondi M, Laukova A, de Niederhausern S, Messi P, Papadopoulou C, Economou V (2020) Controversial aspects displayed by enterococci: probiotics or pathogens? Biomed Res Int. https://doi.org/10.1155/2020/9816185
Perin LM, Miranda RO, Todorov SD, de Melo Franco BDG, Nero LA (2014) Virulence, antibiotic resistance and biogenic amines of bacteriocinogenic lactococci and enterococci isolated from goat milk. Int J Food Microbiol 185:121–126. https://doi.org/10.1016/j.ijfoodmicro.2014.06.001
Park JW, Jeong JS, Lee SI, Kim IH (2016) Effect of dietary supplementation with a probiotic (Enterococcus faecium) on production performance, excreta microflora, ammonia emission, and nutrient utilization in ISA brown laying hens. Poult Sci 95(12):2829–2835. https://doi.org/10.3382/ps/pew241
Preisner O, Lopes JA, Guiomar R, Machado J, Menezes JC (2007) Fourier transform infrared (FT-IR) spectroscopy in bacteriology: towards a reference method for bacteria discrimination. Anal Bioanal Chem 387:1739–1748. https://doi.org/10.1007/s00216-006-0851-1
Ng SC, Hart AL, Kamm MA, Stagg AJ, Knight SC (2009) Mechanisms of action of probiotics: recent advances. Inflamm Bowel Dis 15(2):300–310. https://doi.org/10.1002/ibd.20602
Stout E, Klaenhammer T, Barrangou R (2017) CRISPR-Cas technologies and applications in food bacteria. Annu Rev Food Sci Technol 8:413–437. https://doi.org/10.1146/annurev-food-072816-024723
Barrangou R, Notebaart RA (2019) CRISPR-directed microbiome manipulation across the food supply chain. Trends Microbiol 27(6):489–496. https://doi.org/10.1016/j.tim.2019.03.006
Shokryazdan P, Faseleh Jahromi M, Liang JB, Ho YW (2017) Probiotics: from isolation to application. J Am Coll Nutr 36(8):666–676. https://doi.org/10.1080/07315724.2017.1337529
Prabhurajeshwar C, Chandrakanth RK (2017) Probiotic potential of Lactobacilli with antagonistic activity against pathogenic strains: an in vitro validation for the production of inhibitory substances. Biomed J 40(5):270–283. https://doi.org/10.1016/j.bj.2017.06.008
Funding
This research was supported by Higher Education Commission (HEC; NRPU-20–1339/R&D/09) to Syed Abid Ali.
Author information
Authors and Affiliations
Contributions
Conception, design, and supervision were performed by SAA. Material preparation, data collection, and analysis were performed by AHn, SA, DA, MR, AA, and SAA.
Corresponding author
Ethics declarations
Competing Interests
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Hussain, A., Akram, S., Ahmad, D. et al. Molecular Assessment and Validation of the Selected Enterococcal Strains as Probiotics. Probiotics & Antimicro. Prot. (2023). https://doi.org/10.1007/s12602-023-10163-6
Accepted:
Published:
DOI: https://doi.org/10.1007/s12602-023-10163-6