Abstract
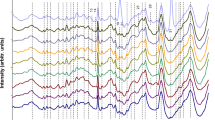
Rapid and reliable discrimination among clinically relevant pathogenic organisms is a crucial task in microbiology. Microorganism resistance to antimicrobial agents increases prevalence of infections. The possibility of Fourier transform infrared (FT-IR) spectroscopy to assess the overall molecular composition of microbial cells in a non-destructive manner is reflected in the specific spectral fingerprints highly typical for different microorganisms. With the objective of using FT-IR spectroscopy for discrimination between diverse microbial species and strains on a routine basis, a wide range of chemometrics techniques need to be applied. Still a major issue in using FT-IR for successful bacteria characterization is the method for spectra pre-processing. We analyzed different spectra pre-processing methods and their impact on the reduction of spectral variability and on the increase of robustness of chemometrics models. Different types of the Enterococcus faecium bacterial strain were classified according to chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis (PFGE). Samples were collected from human patients. Collected FT-IR spectra were used to verify if the same classification was obtained. In order to further optimize bacteria classification we investigated whether a selected combination of the most discriminative spectral regions could improve results. Two different variable selection methods (genetic algorithms (GAs) and bootstrapping) were investigated and their relative merit for bacteria classification is reported by comparing with results obtained using the entire spectra. Discriminant partial least-squares (Di-PLS) models based on corrected spectra showed improved predictive ability up to 40% when compared to equivalent models using the entire spectral range. The uncertainty in estimating scores was reduced by about 50% when compared to models with all wavelengths. Spectral ranges with relevant chemical information for Enterococcus faecium bacteria discrimination were outlined.





Similar content being viewed by others
References
Ausubel F, Brent M, Kingston R, Moore D, Seidman J, Smith J, Struhl K (eds) (1989) Current protocols in molecular biology. Wiley, New York, NY
Brown M, Dunn W, Ellis D, Goodacre R, Handl J, Knowles J, O’Hagan S, Spasic I, Kell D (2005) Metabolomics 1(1):39–51
Neu HC (1992) Science 257:1064–1073
Maquelin K, Kirschner C, Choo-Smith LP, Van den Braak N, Endtz H, Naumann D, Puppels GJ (2002) J Microbiol Methods 51:255–271
Naumann D, Helm D, Labischinski H (1991) Nature 351:81–82
Naumann D, Keller S, Helm D, Schultz Ch, Schrader B (1995) J Mol Struct 347:399–406
Naumann D (2000) Infrared spectroscopy in microbiology. In: Meyers RA (ed) Encyclopedia of analytical chemistry. Wiley, Chichester, pp 102–131
Riddle JW, Kabler PW, Kenner BA, Bordner RH, Rockwood SW, Stevenson HJR (1956) J Bacteriol 72:593–603
Norris KP (1959) J Hyg 57:326–345
Winder CL, Carr E, Goodacre R, Seviour R (2004) J Appl Microbiol 96:328–339
Sandt C, Sockalingum GD, Aubert D, Lepan H, Lepouse C, Jaussaud M, Leon A, Pinon JM, Manfait M, Toubas D (2003) J Clin Microbiol Mar:954–959
Consuelo López-Díez E, Goodacre R (2004) Anal Chem 76:585–591
Wu Q, Hamilton T, Nelson WH, Elliott S, Sperry JF, Wu M (2001) Anal Chem 73:3432–3440
Essendoubia M, Toubasb D, Bouzaggoua M, Pinonb JM, Manfaita M, Sockalingum GD (2005) Biochim Biophys Acta 1724:239–247
Rosch P, Harz M, Peschke K-D, Ronneberger O, Burkhardt H, Popp J (2006) Biopolymers 82:312–316
Xie C, Mace J, Dinno MA, Li YQ, Tang W, Newton RJ, Gemperline PJ (2005) Anal Chem 77:4390–4397
Maquelin K, Kirschner C, Choo-Smith L-P, Ngo-Thi NA,van Vreeswijk T, Stammler M, Endtz HP, Bruining HA, Naumann D, Puppels GJ (2003) J Clin Microbiol Jan:324–329
Goodacre R, Adaoin E, Timmins M, Burton R, Kaderbhai N, Woodward AM, Kell DB, Rooney PJ (1998) Microbiology 144:1157–1170
Mobley P, Kowalski B, Workman J, Bro R (1996) Appl Spectrosc Rev 31:347–368
Udelhoven T, Naumann D, Schmitt J (2000) Appl Spectrosc 54(10):1471–1479(9)
Bakeev KA, Kurtyka B (2005) J Near Infrared Spectrosc 13(6):339–348
Martens H, Nielsen JP, Engelsen SB (2003) Anal Chem 75(3):394–404
Jarvis RM, Goodacre R (2005) Bioinformatics 21(7):860–868
Olivieri AC, Goicoechea HC (2003) J Chemom 17(6):338–345
Geladi P, Kowalsky B (1986) Analytica Chimica Acta 185:1–17
Alsberg B, Kell D, Goodacre R (1998) Anal Chem 70:4126–4133
Martens H, Naes T (eds) (1992) Multivariate calibration. Wiley, UK
Mitchell M (1999) An introduction to genetic algorithms. The MIT Press
Michalewicz Z (ed) (1997) Genetic algorithms+data structures=evolution programs. Springer-Verlag, Berlin Heidelberg New York
Lazraq A, Cléroux R, Gauchi J-P (2003) Chemometr Intell Lab Syst 66:117–126
Graves L, Swaminathan B (2001) Int J Food Microbiol 65:55–62
Brereton RG (ed) (2003) Chemometrics. Data analysis for the laboratory and chemical plant. Wiley
Næs T, Isaksson T, Fearn T, Davies T (eds) (2002) A user-friendly guide to multivariate calibration and classification. NIR Publications, Chichester, p 344
Acknowledgements
O.P. gratefully acknowledges financial support from the Portuguese Foundation for Science and Technology (research grant no. SFRH/BD/15218/2004).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Preisner, O., Lopes, J.A., Guiomar, R. et al. Fourier transform infrared (FT-IR) spectroscopy in bacteriology: towards a reference method for bacteria discrimination. Anal Bioanal Chem 387, 1739–1748 (2007). https://doi.org/10.1007/s00216-006-0851-1
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00216-006-0851-1