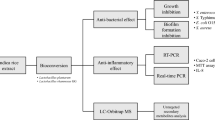
Abstract
Plant-derived lactic acid bacteria are major fermentation organisms that can grow in medicinal herb extracts enriched with phytochemicals like glycosides, phenolic acids, flavonoids, and tannins. Fermentation with strain-specific Lactobacilli harboring metabolic enzymes can increase the bioactivity and bioavailability of medicinal herbs. Fermentation of extracts of Artemisia princeps and Paeonia lactiflora has been previously found to increase their bioactivities. Therefore, this study explores the possibility of increasing the bioactivity of Mentha arvensis (Mentha) extract against lipopolysaccharide (LPS)-induced RAW 264.7 macrophage cells by fermenting with plant-derived probiotic strains Lactobacillus (Lact.) plantarum SN13T and Pediococcus (Ped.) pentosaceus LP28. As a result, fermentation with SN13T significantly increased the bioactivity of Mentha extract as compared to unfermented or LP28-fermented extracts. This higher bioactivity was associated with the metabolism of rosmarinic acid (RA) and caffeic acid (CA), the major bioactive phenolic acids reported in Mentha, along with the production of the metabolite dihydrocaffeic acid (DHCA). DHCA was found to be a more potent LPS-induced nitric oxide (NO) inhibitor than its precursor phenolic acids. The metabolism of RA to DHCA via CA could be mediated by the enzymes cinnamoyl ester hydrolase and hydroxycinnamate reductases, encoded by the ceh gene and the hcrRABC gene operon, respectively, which were identified in the complete genome sequence of Lact. plantarum SN13T but were absent in Ped. pentosaceus LP28. The genes hcrA, hcrB, and hcrC were significantly and time-dependently overexpressed in Lact. plantarum SN13T when grown in the Mentha extract, suggesting the role of phenolic acid metabolism in enhancing its bioactivity.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Lactic acid bacteria (LAB) are the most important microorganisms that are capable of inducing significant changes in the health-promoting properties of plant foods [1]. Plants, particularly, medicinal herbs, are reservoirs of bioactive compounds like glycosides, antioxidants, phenolics, and dietary fibers, as well as vitamins and minerals, which enable species and strain-specific LAB to follow various metabolic routes [2]. During the fermentation process of plants by LAB, the decomposition and/or biotransformation of complex phytochemicals into compatible components can improve their bioavailability and bioactivity, as well as concentrate the functional microbial metabolites with beneficial consequences for human health [3]. LAB species that mainly dominate spontaneous plant fermentation such as Lact. plantarum, Lact. pentosus, and Pediococcus inherit a dedicated portfolio of enzymes like glycosyl hydrolase, phenolic acid decarboxylase and reductase, and esterase activities that catalyze the conversions of glycosides, phenolic acids, and tannins to biologically active metabolites [1].
Over 1200 strains of plant-derived LAB from fruits, vegetables, flowers, and medicinal plants have been isolated and evaluated for their health benefits by our research group. Notably, Lactobacillus plantarum SN13T, which has been isolated from banana leaves, was found to improve liver function by altering the composition of gut microbiota in subjects with mild liver dysfunction [4]. Pediococcus pentosaceus LP28, isolated from the longan fruit Euphoria longana, reduced obesity and fatty liver in high-fat diet-induced mice and reduced BMI, body fat, and waist circumference, suggesting that it is a promising anti-obesity candidate for preventing metabolic syndrome [5]. Furthermore, when Lact. plantarum SN13T or Lact. brevis 174A was grown in medicinal herb extracts like Artemisia princeps Pampanini and Paeonia lactiflora Pall, bioactive compounds were produced that have improved their therapeutic potential [6-8]. Therefore, we speculated that the bioactivity of the medicinal herb, Mentha arvensis Linné var. piperascens Malinvaud (Lamiaceae), i.e., Mentha, can also be enhanced by fermentation with plant-derived LAB.
Mentha, among the most popular herbs, is widely used in cooking and cosmetics. Traditionally, it has been used for the treatment of gastrointestinal disorders such as flatulence, indigestion, nausea, vomiting, anorexia, and ulcerative colitis. Moreover, the essential oil and extracts of Mentha species have been reported to possess antimicrobial, fungicidal, antiviral, insecticidal, and antioxidant properties [9]. Apart from essential oils such as menthol, the genus Mentha is known to be rich in phenolic compounds, including rosmarinic acid and caffeic acid, its major bioactive chemical constituents [9-11]. While few studies have already reported the bioactivities of fermented Mentha extract [12-14], a more detailed study focusing on bacterial strain-specific changes in metabolites and gene expression of metabolic enzymes involved in the fermentation of Mentha extract is yet to be explored. Thus, in this work, we report the increased bioactivity of the Mentha extract against LPS-stimulated RAW 264.7 cells by fermentation with strain-specific LAB, along with the changes in major phenolic acid concentrations, the detection of a newly produced bioactive metabolite, and the identification and overexpression of phenolic acid metabolism-related genes in L. plantarum SN13T during the fermentation.
Materials and Methods
Bacteria Culture and Fermentation Conditions
The lactic acid bacterial strains Lactobacillus plantarum SN13T and Pediococcus pentosaceus LP28, which have been isolated previously from plant sources—banana leaves and longan fruit Euphoria longana, respectively—were grown at 37 °C overnight in MRS broth (Merck, Germany). After cultivation, the bacterial cells were collected by centrifugation at 8000 × g for 10 min.
Mentha arvensis Linné var. piperascens Malinvaud (Lamiaceae), i.e., Mentha herb (5 g), purchased from Kojima Kampo Co., Ltd. (Osaka, Japan), was extracted by suspending and boiling in 100 ml of distilled water for 30 min. After cooling to room temperature, it was centrifuged at 5000 × g for 10 min and filtrated with a 0.22 µm membrane filter (Advantec Ltd., Japan) to obtain the aqueous extract of Mentha arvensis (MA). Then, the overnight bacterial cells (approximately 3 × 109 colony forming units/ml) obtained by centrifugation were inoculated into the MA extract and incubated at 30 °C with shaking at 120 rpm. These conditions were chosen on the basis of previous literatures of plant-food fermentations where hydroxycinnamic acids (like rosmarinic acid and caffeic acid in Mentha extract) are abundant [15, 16]. After 24 h (h), the pH dropped to 4.4 in the fermented extracts (fMA-SN13T and fMA-LP28), and they were finally collected by centrifugation at 5000 × g for 10 min and subsequent filtration.
For the RT-qPCR experiments of bacterial phenolic acid metabolism genes, the overnight cells of Lact. plantarum SN13T were incubated with MRS, MRS supplemented with 1 mg/ml of rosmarinic acid (MRS + RA), or MA for 5 h or 24 h at 30 °C with shaking at 120 rpm.
Cell Culture and Treatment
Murine macrophage-like cell line RAW 264.7 cells (RRID: CVCL_0493) were grown in DMEM medium supplemented with 10% FBS and 100 µg/ml of penicillin/streptomycin by incubation in a humidified 5% CO2 atmosphere at 37 °C, as described previously [7]. To stimulate the cells, the medium was exchanged with fresh DMEM medium with 0.5% FBS, and 1 µg/ml LPS was added in the presence or absence of the extracts (0.5% to 1% (v/v) final concentration) or standard solutions of rosmarinic acid (RA), caffeic acid (CA), or dihydrocaffeic acid (DHCA) at concentrations of 5 µg/ml to 60 µg/ml and incubated for 5 h or 24 h.
Cell Viability Assay
Cell viability was determined using a CCK-8 kit (Dojindo, Japan) in accordance with the manufacturer’s instructions. Briefly, 100 μl of cells at a density of 1.8 × 105 cells per well was incubated with LPS (1 µg/ml) in the presence or absence of the extracts or standard solutions for 24 h. CCK-8 solution (10 μl) was added to each well, and the cells were incubated for another 2 h. Then, the absorbance at 450 nm was measured. The percentage of viable cells was determined as a value relative to untreated cells.
Measurement of Intracellular ROS Levels
The RAW 264.7 macrophage cells were treated with LPS in the presence or absence of the extracts or standard solutions for 24 h. After incubating the cells with 10 mM DCFH-DA for 30 min at 37 °C and washing twice with phosphate-buffered saline, the DCF fluorescence was measured at excitation and emission wavelengths of 485 and 530 nm, respectively [17]. The Relative Fluorescence Unit (RFU) thus obtained represented the intracellular ROS levels.
Measurement of NO Production
The nitric oxide (NO) levels were determined by measuring the nitrite in the supernatant from the cell culture treated with LPS for 24 h. Equal volumes of the cell culture supernatants and the Griess reagent (1% sulfanilamide in 5% phosphoric acid and 0.2% naphthyl ethylenediamine dihydrochloride) were mixed. After 5 min at room temperature, the absorbance at 550 nm was measured. The NO concentration was calculated from the standard curve of sodium nitrite [18].
Inflammatory Cytokine Determination
The cells were treated with LPS in the presence or absence of the extracts or standard solutions for 24 h. The concentrations of inflammatory cytokines IL-1β, IL-6, and TNF-α in the cell supernatants were determined by respective ELISA kits (BioLegend, USA), following the manufacturer’s protocols.
RNA Extraction and qRT-PCR Analysis
RNA extraction and qRT-PCR were performed separately for samples treated with RAW 264.7 cells and bacterial cells grown in MRS and MA, to study the gene expression of LPS-stimulated inflammatory mediators and the gene expression of phenolic acid metabolism genes, respectively.
The total RNA from RAW 264.7 cells treated for 5 h with the extracts or standard solutions and Lact. plantarum SN13T cells grown in MRS, MRS-RA, and MA for 5 h or 24 h were isolated using NucleoSpin RNA plus (Macherey–Nagel GmbH and Co. KG, Germany). ReverTra Ace qPCR RT Master Mix with gDNA remover (Toyobo, Japan) was used for gDNA removal and reverse transcription in accordance with the manufacturer’s instruction manual. qRT-PCR was conducted on the CFX Maestro 2.3 real-time PCR system (Bio-Rad, USA) using the KAPA SYBR Fast qPCR Kit (Kapa Biosystems, USA). The qPCR was conducted under the following conditions: an initial 2 min at 95 °C, followed by 40 cycles of 5 s at 95 °C and 10 s at 60 °C. The relative transcriptional levels were normalized to housekeeping genes, i.e., gapdh for RAW 264.7 cells and ldh for bacterial cells. The gene expressions were analyzed using the ΔΔCT method. As given in Table 1, the primers used for qRT-PCR of LPS-induced inflammatory genes of RAW 264.7 cells were reported in a previous study [18], while the primers of SN13T genes were designed using Primer-BLAST online tool [19]. All qPCR assays amplified a single product, as determined by melting curve analysis and shown in Supplementary Figs. 1 and 2.
Identification and Determination of Metabolites in Unfermented and Fermented Mentha Extract
HPLC analyses were performed to compare the constituents of unfermented and fermented Mentha extracts according to a previous study with modifications [7]. Aliquots (2.5 µl) of the MA, fMA-SN13T, and fMA-LP28 extracts were applied to HPLC (JASCO system; JASCO Corporation) with a YMC-Pack ODS-AQ (150 × 4.6 mm, 5 µm, 12 nm) column (YMC, Japan). The column was equilibrated with water containing 0.1% trifluoracetic acid, and gradient elution was performed with 0% to 10%, 10 to 40%, and finally 40 to 60% acetonitrile over 20 min, 30 min, and 10 min sequentially at a flow rate of 1 ml/min. The elution profiles were monitored at absorbances of 320 nm and 280 nm. The chromatogram of the MA was compared with those of the fermented extracts, fMA-SN13T and fMA-LP28. Rosmarinic acid and caffeic acid in all of the extracts were identified using the respective analytical standards, and their concentrations were determined by standard curves at the absorbance of 320 nm. At 280 nm, a newly produced compound was detected in fMA-SN13T. The compound was purified from fMA-SN13T by ethyl acetate extraction, followed by preparative TLC using a mobile phase of methanol: chloroform: toluene at 5:4:1, and followed by HPLC.
The purified compound was identified using a combination of GC–MS as well as 1H-NMR and 13C-NMR spectra. The MS spectra were captured on a Thermo Fisher Scientific LTQ Orbitrap XL. GC–MS was performed on a JMS-T100GCV AccuTOF GCv4G (JEOL Ltd.) with an HP-5MS capillary column (0.25 mm × 0.25 µm × 30 m). The parameters for EI mode were an ion-source temperature of 250 °C, an electron energy of 70 eV, and a filament current of 300 µA. 1H-NMR and 13C-NMR spectra were captured on a JEOL JNM-LA500 spectrometer at 600 MHz. Finally, the identified compound was confirmed by HPLC using the analytical standard.
Sequence Data Analyses
Homology searches were performed on the whole genome sequences of Lact. plantarum SN13T and Ped. pentosaceus LP28 using the software in silico Molecular Cloning (R) Genomics Edition version 6.0.30D and the BLAST algorithm on the National Center for Biotechnology Information server (https://blast.ncbi.nlm.nih.gov/Blast.cgi) [20], respectively. Multiple alignments were made using the Clustal Omega program (https://ebi.ac.uk/Tools/msa/clustalo/) on the EBI site after the retrieval of sequences from the GenBank database [21].
Statistical Analyses
All data were presented as the means and SD of triplicates. The significance of differences was determined via ANOVA, followed by a post hoc Tukey test, and differences with p < 0.05 were considered statistically significant.
Results
Increased Bioactivity of MA Extract by Fermentation with Lact. plantarum SN13T
When RAW 264.7 cells were stimulated with LPS (1 µg/ml) for 24 h, the levels of inflammatory mediators NO, ROS, IL-1β, IL-6, and TNF-α in cell supernatants were found to be significantly more increased than those in untreated cells, as seen in Fig. 1a–c. At a 0.5% or 1% concentration, all of the extracts displayed cell viability similar to that of control cells. Figure 1a shows that, in the presence of MA or fermented MA at 0.5% or 1% final concentrations, the LPS-induced NO levels were significantly decreased. At 0.5%, significant decreases in NO as compared to the unfermented MA were observed in fMA-SN13T but not in fMA-LP28. Similar results are seen in Fig. 1b, where all of the extracts—MA, fMA-SN13T, and fMA-LP28—could significantly decrease the LPS-induced intracellular ROS levels at both concentrations. The highest decrease was shown in fMA-SN13T, although it was not significantly higher than in other extracts. Meanwhile, Fig. 1c depicts significant decreases in the levels of LPS-induced cytokines IL-1β, IL-6, and TNF-α after treatment with MA or fermented MA at a 1% concentration. Here, as well, fMA-SN13T was found to be the most bioactive of the extracts.
Increased bioactivity of MA extract by fermentation with strain-specific Lactobacillus strains. a Nitric oxide level (µM) in LPS-stimulated RAW 264.7 cells treated with the extract MA, fMA-SN13T, or fMA-LP28, expressed in comparison with the control. b Intracellular ROS levels (RFU) in LPS-stimulated RAW 264.7 cells treated with MA, fMA-SN13T, or fMA-LP28, expressed in comparison with the control. c Cytokine (IL-1β, TNF-α, and IL-6) levels (pg/ml) in LPS-stimulated RAW 264.7 cells treated with MA, fMA-SN13T, or fMA-LP28, expressed in comparison with the control. d Relative mRNA expressions of iNOS, sod-2, il-1β, tnf-α, and il-6 in LPS-stimulated RAW 264.7 cells treated with MA, fMA-SN13T, or fMA-LP28. All data are expressed as the mean value of triplicate experiments. Error bars represent ± standard deviation. *p < 0.05 versus LPS and #p < 0.05 versus MA-treated cells
To confirm the bioactivity of MA and fermented MA extracts, the relative gene expression levels of iNOS, sod-2, il-1β, il-6, and tnf-α in RAW 264.7 cells stimulated with LPS for 5 h were determined in the presence of the extracts MA, fMA-SN13T, and fMA-LP28 at a 0.5% concentration, as shown in Fig. 1d. While the relative gene expression of inflammation-related NO-producing enzyme iNOS was significantly downregulated in the presence of MA and fMA-SN13T, it was notable that its expression in the presence of fMA-SN13T was seen to be significantly reduced even as compared to that of MA. However, the treatment of fMA-LP28 could not significantly reduce the iNOS mRNA expression as compared to that of LPS-treated cells. Consistent with the results of the previous experiment, the fermented or unfermented MA could decrease the LPS-induced gene expressions of inflammatory cytokines il-1β, il-6, and tnf-α and antioxidant enzyme sod-2. In these cases, the fermented extract, fMA-SN13T, had higher activity than MA, while fMA-LP28 extract demonstrated the least activity.
Metabolism of Rosmarinic Acid (RA) in MA by Lact. plantarum SN13T
We performed HPLC analyses of the extracts MA, fMA-SN13T, and fMA-LP28 to compare the chemical constituents in the MA extract after fermentation. In Fig. 2a, the HPLC chromatograms of the extracts detected at the absorbance of 320 nm show peaks of rosmarinic acid and caffeic acid at retention times of 39.52 min and 27.18 min, respectively. It was observed that the concentration of rosmarinic acid significantly decreased in fMA-SN13T as compared to that in unfermented MA—from 314.45 µg/ml to 192 µg/ml—while only an insignificant decrease in the fMA-LP28 extract, and no significant change in the caffeic acid concentration was measured among the three extracts as shown in Fig. 2b.
a HPLC chromatograms of MA, fMA-SN13T, and fMA-LP28 extracts and the analytical standards of rosmarinic acid and caffeic acid at the absorbance of 320 nm. The peaks enclosed in boxes with gray dashes and black dots represent rosmarinic acid and caffeic acid, respectively. b Concentrations (µg/ml) of rosmarinic acid and caffeic acid measured in MA, fMA-SN13, and fMA-LP28 extracts as determined from HPLC standard curves. Data are expressed as the mean value of triplicate experiments. Error bars represent ± standard deviation. ***p < 0.01 versus MA
Identification of a Newly Produced Compound in the Fermented Extract
On further analyses of the HPLC chromatograms of the extracts MA, fMA-SN13T, and fMA-LP28 at the absorbance of 280 nm, we detected a newly produced peak at the retention time of 23.6 min in the fMA-SN13T extract. However, it was undetectable in both unfermented MA and fMA-LP28, as shown in Fig. 3a. The accumulation of this metabolite in fMA-SN13T could be responsible for the higher bioactivity than in the other extracts. Therefore, we purified this compound from the extract fMA-SN13T sequentially by organic solvent extract, preparative TLC, and HPLC. For the identification, this purified compound was analyzed for GC–MS, 1H-NMR, and 13C-NMR spectra, as shown in Figs. 3b, S3a, and S3b, respectively. The number of molecular ions of the underivatized compounds detected as [M–H]− in the negative ion ESI–MS spectra was determined to be 182, with the molecular formula C9H10O4. The GC–MS data showed three TMS groups on the derivatized sample with a calculated M+ of 398 and fragment ions (m/z) of 383, 282, 267, and 179. The δC (chemical shifts in ppm) in 13C-NMR spectra were observed at 179.957, 146.203, 144.604, 133.746, 120.447, 116.339, 37.156, and 31.469. Taken together, the NMR data and MW data were consistent with previous reports of the 2,3-dihydro derivative of caffeic acid [22]. Finally, after confirming the purified peak to be 3-(3′,4′-dihydroxyphenyl) propanoic acid or dihydrocaffeic acid (DHCA) by comparing the HPLC chromatogram of its analytical standard, we determined its concentration produced in fMA-SN13T to be 71.78 µg/ml. The possible metabolic pathway, by which the excess caffeic acid in the MA extract fermented with Lact. plantarum SN13T formed by the hydrolysis of rosmarinic acid via bacterial esterases is further reduced into dihydrocaffeic acid via reductases, is depicted in Fig. 3c.
a HPLC chromatograms of MA, fMA-SN13T, and fMA-LP28 extracts and analytical standards of dihydrocaffeic acid at the absorbance of 280 nm. b GC–MS spectra (EI mode) of purified dihydrocaffeic acid derivatized with TMS. c Possible metabolic pathway of dihydrocaffeic acid from rosmarinic acid via caffeic acid by the actions of bacterial esterase and reductase
Dihydrocaffeic Acid is a More Potent NO Inhibitor Than Caffeic Acid and Rosmarinic Acid
We compared the activities of RA, CA, and DHCA in various concentrations in our experimental model of LPS-induced RAW 264.7 cells. All three had decreased cell viability above a concentration of 80 µg/ml; thus, we determined the NO and intracellular ROS levels of LPS-induced cells when treated with concentrations of 5 µg/ml, 10 µg/ml, 20 µg/ml, 40 µg/ml, and 60 µg/ml. As seen in Fig. 4a, at the highest concentration of 60 µg/ml, all RA, CA, and DHCA could significantly lower the LPS-induced NO in RAW 264.7 cells. At lower concentrations, from 5 µg/ml to 20 µg/ml, DHCA markedly decreased the NO levels as compared to both CA and RA; i.e., DHCA was the most potent inhibitor of LPS-induced NO. Similarly, Fig. 4b shows that CA and DHCA decreased the LPS-induced intracellular ROS significantly as compared to RA at concentrations of 5 µg/ml, 40 µg/ml, and 60 µg/ml. In addition, all RA, CA, and DHCA at a concentration of 20 µg/ml significantly decreased the mRNA expression of iNOS as compared to the LPS-only-treated cells, but any significant differences between the sample groups were not observed in the case of the antioxidant enzyme sod-2 gene and proinflammatory cytokine il-1β, tnf-α, and il-6 genes, as shown in Fig. 4c.
a Effect of rosmarinic acid (RA), caffeic acid (CA), and dihydrocaffeic acid (DHCA) on NO levels (µM) produced by LPS-induced RAW 264.7 cells at concentrations of 5 µg/ml, 10 µg/ml, 20 µg/ml, 40 µg/ml, and 60 µg/ml. b Effects of RA, CA, and DHCA on intracellular ROS levels (RFU) produced by LPS-induced RAW 264.7 cells at concentrations of 5 µg/ml, 10 µg/ml, 20 µg/ml, 40 µg/ml, and 60 µg/ml. c Effect of RA, CA, and DHCA on relative mRNA expressions of iNOS, sod-2, il-1β, tnf-α, and il-6 in LPS-induced RAW 264.7 cells at a concentration of 20 µg/ml. Data are expressed as the mean value of triplicate experiments. Error bars represent ± standard deviation. *p < 0.05 versus LPS and #p < 0.05 versus DHCA-treated cells
Identification of Phenolic Acid Metabolism-Related Genes in Lact. plantarum SN13T
On analysis of the complete genome sequences, AP019815.1 and NZ_DF970691.1 of Lact. plantarum SN13T and Ped. pentosaceus LP28, respectively, for annotated cinnamoyl ester hydrolase (ceh), none was found in strain LP28, while a putative enzyme with 114 amino acids was found in SN13T (SN13T_1651), showing 35% similarity, as given in Table 2, and aligned with ceh homolog Lj0536 in Lact. johnsonii N6.2 in Fig. S4a. As shown in Table 2, SN13T harbored the complete hcr operon, consisting of a Lys-R type transcriptional regulator, NADPH-dependent FMN reductase family protein, NADPH-dependent FMN reductase, and a hypothetical protein encoded by SN13T_0395, SN13T_0394, SN13T_0393, and SN13T_0392 that exhibited only 86%, 94%, 90%, and 85% amino acid identity to hcrR, hcrA, hcrB, and hcrC of Lact. plantarum WCFS1, respectively (see Figure S4b-e, respectively).
Overexpression of Phenolic Acid Metabolism-Related Genes in Lact. plantarum SN13T During MA Fermentation
After the identification of phenolic acid metabolism-related genes in Lact. plantarum strain SN13T, we speculated that these genes were overexpressed during the fermentation of the plant extract MA. Hence, we compared the expressions of genes ceh, hcrR, hcrA, hcrB, and hcrC when these strains were grown in MRS, MRS supplemented with RA at 1 µg/ml, and MA for 5 h. As shown in Fig. 5a, the esterase gene ceh was significantly upregulated in the presence of RA, as well as the MA extract, as compared with MRS broth. In MA, the relative expression of hcrB reached higher than 250-fold as compared to MRS, along with the significant overexpression of the remaining hcr operon. Additionally, it was found that the esterase gene ceh and all genes hcrRABC of reductase operon were increasingly upregulated at 5 h and 24 h of growth, as shown in Fig. 5b, which supports that phenolic acid-metabolism genes of Lact. plantarum SN13T are time-dependently overexpressed, producing DHCA in the MA extract during the 24 h fermentation.
a Relative mRNA expressions of phenolic metabolism-encoding genes ceh, hcrR, hcrA, hcrB, and hcrC, when Lact. plantarum SN13T was grown in MRS, MRS with rosmarinic acid (1 µg/ml), and MA for 5 h. b Relative mRNA expressions of phenolic metabolism–encoding genes ceh, hcrR, hcrA, hcrB, and hcrC, when Lact. plantarum SN13T was grown in MA for 0 h, 5 h, or 24 h. Data are expressed as the mean value of triplicate experiments. Error bars represent ± standard deviation. *p < 0.05, **p < 0.01, and ***p < 0.001 versus MRS or 0 h
Discussion
Our results demonstrated that fermentation with Lact. plantarum SN13T could enhance the potency of the MA extract, while fermentation with Ped. pentosaceus LP28 did not show significantly better bioactive properties in the in vitro experimental model of LPS-stimulated RAW 264.7 murine macrophages. An anti-inflammatory drug, dexamethasone at 0.5 µg/ml, also significantly reduced NO concentration and iNOS expression in the same in-vitro model in our previous unpublished study, which is comparable to the bioactivity of unfermented or SN13T fermented Mentha extracts (see Supplemental Figure S5). The LPS-induced RAW 264.7 macrophage model was also used to study the anti-inflammatory effect of the stem extract of Alternanthera sessilis, where its potential to suppress the proinflammatory mediators and cytokines was compared to dexamethasone [23]. In our previous study, fermentation with Lact. brevis 174A strain enhanced the antioxidant and anti-inflammatory activities of the medicinal herb extract Paeonia Radix Alba by increasing the total phenolic content (TPC) and producing the gallic acid metabolite, pyrogallol [7]. Numerous other reports have also shown that changes in the phenolic content are associated with simultaneous changes in antioxidant activities after Lactobacillus fermentation of various food matrices, due to the enzymatic actions of any of the commonly inherited enzymes of Lactobacillus species, such as glucosidase, esterase, phenolic acid decarboxylase, phenolic acid reductase, or tannase [24-29]. In addition, it has been well reported that the ability of LAB to improve the bioavailability and bioactivity of phytochemicals is associated with species- or strain-specific metabolic features [3]. The strain Lact. plantarum SN13T has been previously found to produce IL-8-inhibiting metabolites like catechol and secotanapartholide C in the medicinal herb extract of Artemisia princeps Pampanini. It was shown that this strain could grow vigorously in different herbal extracts, as it harbors a β-glucosidase enzyme encoding 11 open reading frames (ORFs) [6]. Thus, we speculated that, when grown in the Mentha herb, Lact. plantarum SN13T also produces bioactive metabolites that potentiate its bioactivity as compared to other strains.
In the genus Mentha, rosmarinic acid and caffeic acid are reported to be the most important bioactive phenolic acid metabolites [30]. In fact, rosmarinic acid is one of the marker compounds used to evaluate the quality of M. arvensis L. [31]. Thus, the significant decrease in RA concentration after SN13T fermentation of MA was a notable observation as it could be co-related with the significantly increased bioactivity of fMA-SN13T extract. However, it has been reported that M. arvensis L. extract fermented with a combination of Lact. rhamnosus, E. faecium, and Lact. acidophilus showed an increase in the concentration of rosmarinic acid that could suppress MDA, NO, and corticosterone; improve body weight; decrease daily food intake and duodenum histology; and increase serotonin and β-endorphin levels in immobilization-induced stressed rats [12]. In another study, such fermented M. arvensis extract administration also provided neuroprotection against transient global cerebral ischemia in gerbils and SH-SH5Y cells by downregulating MAPK signaling [13]. In addition, M. piperita fermented with Bacillus subtilis suppressed PMA-induced ROS, ERK, and MUC5AC mRNA protein expression in lung epithelial cells [14]. Nonetheless, these previous reports confirm the functional aspects of the strain-specific fermentation of Mentha herb extracts, but without a clear understanding of the metabolite profiling of such fermented extracts.
Meanwhile, there have been several reports suggesting that rosmarinic acid can be readily degraded into caffeic acid and derivatives by gut microbiota in vitro as well as after oral intake in mice and humans [32, 33]. Lact. johnsonii was reported to metabolize RA from rosemary extract with cinnamoyl esterases into caffeic acid and 3,4-dihyroxyphenyllactic acid in vitro and in a gastrointestinal model [34]. The microbial catabolism of RA was also confirmed in thyme phenol-enriched olive oil, yielding hydroxyphenylpropionic acid as the main metabolite via CA [35]. RA was also reported to be degraded into conjugated forms of caffeic acid, ferulic acid, and m-coumaric acid following intake of Perilla frutescens [36]. While the immediate metabolite of RA has been consistently identified as CA in several reports [30, 34, 35, 37], CA is known to be either reduced into dihydrocaffeic acid or decarboxylated into vinyl catechol via microbial phenolic acid reductase or decarboxylase, respectively [38, 39]. Other studies have reported the final metabolite of CA to be 3-hydroxyphenylpropionic acid (3-HPP) or 4-ethylcatechol via further degradation [35, 40, 41]. Since there was no significant change in the CA concentration among the three extracts in our study, this led us to hypothesize that the CA produced from rosmarinic acid was further degraded into other bioactive compounds in fMA-SN13T. In our results, only DHCA was detected as a phenolic acid metabolite in the SN13T-fermented MA. Strain-specific metabolism of CA during in vitro Lactobacillus fermentation of phenolic-rich plant foods such as cherry juice, broccoli puree, and elderberry juice also produced DHCA through phenolic acid decarboxylases and reductases [42, 43].
To explore the possibility of the increased bioactivity of the fMA-SN13T extract as compared to unfermented MA and fMA-LP28 extracts to be associated with the concentration of DHCA, we studied its bioactivity with respect to its metabolic precursors, RA and CA. RA, CA, and caffeoyl derivatives have been reported to have in vitro antioxidant activity to scavenge superoxide and hydroxyl radicals and a potent anti-inflammatory activity resulting from decreased arachidonate formation in previous studies [44, 45]. Our results suggest that the bioactivity of the phenolic acid metabolites against LPS-induced RAW 264.7 cells could be in the increasing order of RA, CA, and DHCA. Although DHCA is not one of the most commonly found hydroxycinnamic acids, its precursors, such as feruloyl podospermic acid, catechins, procyanidins, and caffeic acid, are found in different food sources [46]. It has been determined to be one of the major phenolic acids found in human fecal water, blood, and urine as a metabolite after the intake of various polyphenols present in food, beverages, and medicinal plants or extracts—such as coffee, artichoke leaf extracts, and chocolate—and in rat urine after the ingestion of polyphenol-rich wine extract [47-50]. Similar to our results, other studies have reported DHCA to be the most potent anti-DPPH (2,2-diphenyl-1-picrylhydrazyl radical) compound among CA, DHCA, and their corresponding n-alkyl esters [51]. DHCA, along with the related colonic metabolite dihydroferulic acid, was found to be more effective inhibitors of in vitro platelet activation than their phenolic precursors present in green coffee bean extract and yerba mate extract via P-selectin expression, suggesting an increase in efficacy with the metabolism of phenolic compounds [52]. DHCA can also function as an intracellular antioxidant in human EA.hy926 epithelial cells by increasing eNOS activity [53]. It reduces the cytotoxicity and pro-inflammatory cytokine IL-6 and IL-8 production in UV-irradiated HaCaT keratinocyte cells, resulting from the combined effect of direct radical scavenging of the ROS or reinforcement of the antioxidant potential of the keratinocytes, and a direct interference with the cytokine signaling pathway [46]. Recently, a high-throughput screening identified DHCA as being one of the two phytochemicals effective in promoting resilience against stress via reducing IL-6 production by inhibiting DNA methylation, which modulated the brain synaptic plasticity and peripheral inflammation, making it a strong candidate for treating stress disorders and depression, either alone or in combination with currently available antidepressants [54].
Lactobacillaceae are known to possess a broad spectrum of enzymes—such as esterases, reductases, and decarboxylases—for the biotransformation of bioactive dietary phenolic compounds such as hydroxycinnamic acids and hydroxybenzoic acids [39]. Several Lactobacillus strains have been characterized for their cinnamoyl esterase activity; however, such esterases—Lj0536 and Lj1228, reported to have substrate specificity for RA—have only been characterized for Lact. johnsonii N6.2 [37]. The ceh gene annotated in the whole genome sequence of Lact. plantarum SN13T encodes an alpha/beta fold hydrolase family enzyme, displaying no significant similarities with previously reported esterases Lj1228, HceP, Lp_0796, and Est_1092 from Lact. johnsonii N6.2, Lact. plantarum TMW1.46, Lact. plantarum WCFS1, and Lact. plantarum DSM 1055, respectively [39]. These findings reinforce that the characterization of the substrate specificity of hydroxycinnamic acid esterases is limited to the use of a few model compounds and does not reflect the diversity of phenolic acid esters in plants and even in esterases in Lact. plantarum species.
On the other hand, the gene cluster involved in hydroxycinnamate reduction, hcrAB, was identified and characterized recently in Lact. plantarum WCFS1 [15]. The inducible reductase activity was not widely present among lactic acid bacteria and was reportedly 100 times lower than decarboxylase activity [55]. The presence of a functional hcrB gene was reported to be a minimum criterion for hydroxycinnamate reductase activity in Lact. plantarum [16]. Unlike in Ped. pentosaceus LP 28, putative phenolic acid reductases were identified in the complete genome of Lact. plantarum strains SN13T homologous to hcrB in Lact. plantarum WCFS1, along with other proteins that were encoded in the proposed operon, including hcrR, hcrA, and hcrC. Homologs of hcrB were also identified in Lact. rossiae (Par1) and Lact. fermentum (hcrF) having 25% and 63% amino acid identity, respectively [16]. In the same study, phenolic acid reductases were not identified in any of the Pediococcus strains, which is consistent with our report. It has been suggested that heterofermentative lactic acid bacteria use hydroxycinnamic acids as external acceptors of electrons to gain additional metabolic energy to combat the stressful conditions generated by phenolic acids [38]. In turn, this metabolic adaptive feature might be responsible for increasing the bioactivity and bioavailability of dietary phenolic acids.
The differential relative gene expressions of hcrR, hcrA, hcrB, and hcrC have been previously reported for Lact. plantarum WCFS1 when exposed to different hydroxycinnamic acids, such as p-coumaric acid, m-coumaric acid, o-coumaric acid, ferulic acid, caffeic acid, and sinapic acid [15]. These genes were also induced in the presence of a nonhydroxy-derived cinnamic acid, possibly, due to structural similarity, but not in the presence of hydroxybenzoic acids such as gallic acid [15]. Although slight similarity of the amino acid in the reductase enzymes was observed, the patterns of relative expression of hcrR, hcrA, hcrB, and hcrC genes by Lact. plantarum SN13T in the presence of RA or MA were found to be different from those of Lact. plantarum WCFS1 when exposed to caffeic acid [15]. The biotransformation of phenolic compounds by lactic acid bacteria does not always correlate with the presence or absence of enzymes and the metabolic activity in laboratory media [39], which may be associated with differential enzyme gene expressions in the presence of complex substrates. Such strain-specific expressions of genes encoding phenolic metabolism and their substrate-based differential regulation were also reported in olive extract, millet, and sorghum fermentations [56, 57]. Hence, our results support the notion that strain-specific enzymes metabolizing phenolic acids, the differential expression of genes encoding-related enzymes, and unlimited variation in composition of individual fermentation sources—foods, medicinal extracts, or media—ultimately affect the behavior of fermenting microbiota and their functional aspects.
To summarize, the fermentation of the herb Mentha extract with Lact. plantarum SN13T increases its antioxidant and anti-inflammatory bioactivity against LPS-induced macrophage cells. This effect was associated with the metabolism of RA into possibly potent metabolite DHCA via the overexpression of microbial strain-specific phenolic metabolism genes, i.e., esterase ceh and reductases hcrR, hcrA, hcrB, and hcrC in Mentha extract fermentation. Hence, we conclude that fermenting medicinal herbal extracts with specific plant-derived LAB strains could be a significant technique for enhancing their therapeutic potential.
Data Availability
All datasets generated for this study are included in the article/Supplementary Material.
References
Filannino P, Di Cagno R, Gobbetti M (2018) Metabolic and functional paths of lactic acid bacteria in plant foods: get out of the labyrinth. Curr Opin Biotechnol 49. https://doi.org/10.1016/j.copbio.2017.07.016
Di Cagno R, Filannino P, Gobbetti M (2016) Novel fermented fruit and vegetable-based products. In: Food Eng Ser
Hussain A, Bose S, Wang J-H et al (2016) Fermentation, a feasible strategy for enhancing bioactivity of herbal medicines. Food Res Int 81. https://doi.org/10.1016/j.foodres.2015.12.026
Higashikawa F, Danshiitsoodol N, Kanno K et al (2020) Lactobacillus plantarum SN13T cells improve hepatic dysfunction and fecal microbiota: a randomized pilot study. Arch Clin Biomed Res 04. https://doi.org/10.26502/acbr.50170129
Higashikawa F, Noda M, Awaya T et al (2016) Antiobesity effect of Pediococcus pentosaceus LP28 on overweight subjects: a randomized, double-blind, placebo-controlled clinical trial. Eur J Clin Nutr 70:582–587. https://doi.org/10.1038/ejcn.2016.17
Okamoto T, Sugimoto S, Noda M, et al (2020) Interleukin-8 release inhibitors generated by fermentation of Artemisia princeps Pampanini herb extract with Lactobacillus plantarum SN13T. Front Microbiol 11. https://doi.org/10.3389/fmicb.2020.01159
Shakya S, Danshiitsoodol N, Sugimoto S et al (2021) Anti-oxidant and anti-inflammatory substance generated newly in Paeonia radix Alba extract fermented with plant-derived Lactobacillus brevis 174A. Antioxidants 10. https://doi.org/10.3390/antiox10071071
Shakya S, Danshiitsoodol N, Noda M et al (2022) 3-Phenyllactic acid generated in medicinal plant extracts fermented with plant-derived lactic acid bacteria inhibits the biofilm synthesis of Aggregatibacter actinomycetemcomitans. Front Microbiol 13. https://doi.org/10.3389/fmicb.2022.991144
Fatih B, Madani K, Chibane M, Duez P (2017) Chemical composition and biological activities of Mentha species. In: Aromatic and Medicinal Plants - Back to Nature. InTech
Bodalska A, Kowalczyk A, Włodarczyk M, Fecka I (2019) Analysis of polyphenolic composition of a herbal medicinal product—peppermint tincture. Molecules 25:69. https://doi.org/10.3390/molecules25010069
Xu L-L, Xu J-J, Zhong K-R et al (2017) Analysis of non-volatile chemical constituents of Menthae Haplocalycis Herba by ultra-high performance liquid chromatography-high resolution mass spectrometry. Molecules 22:1756. https://doi.org/10.3390/molecules22101756
Tian W, Akanda M, Islam A et al (2018) The anti-stress effect of Mentha arvensis in immobilized rats. Int J Mol Sci 19:355. https://doi.org/10.3390/ijms19020355
Islam MS, Shin H-Y, Yoo Y-J et al (2022) Fermented Mentha arvensis administration provides neuroprotection against transient global cerebral ischemia in gerbils and SH-SY5Y cells via downregulation of the MAPK signaling pathway. BMC Complement Med Ther 22:172. https://doi.org/10.1186/s12906-022-03653-7
Jee HJ, Bormate KJ, Lim O, Jung Y-S (2021) Inhibitory effect of fermented Mentha piperita on MUC5AC in lung epithelial cells. J Korean Soc Food Sci Nutr 50:330–338. https://doi.org/10.3746/jkfn.2021.50.4.330
Santamaría L, Reverón I, López de Felipe F et al (2018) Unravelling the reduction pathway as an alternative metabolic route to hydroxycinnamate decarboxylation in Lactobacillus plantarum. Appl Environ Microbiol 84. https://doi.org/10.1128/AEM.01123-18
Gaur G, Oh J-H, Filannino P et al (2020) Genetic determinants of hydroxycinnamic acid metabolism in heterofermentative Lactobacilli. Appl Environ Microbiol 86. https://doi.org/10.1128/AEM.02461-19
Li P, Li Z (2015) Neuroprotective effect of paeoniflorin on H2O2-induced apoptosis in PC12 cells by modulation of reactive oxygen species and the inflammatory response. Exp Ther Med 9. https://doi.org/10.3892/etm.2015.2360
Hwang J, Ma J, Park J et al (2018) Anti-inflammatory and antioxidant effects of MOK, a polyherbal extract, on lipopolysaccharide-stimulated RAW 264.7 macrophages. Int J Mol Med. https://doi.org/10.3892/ijmm.2018.3937
Ye J, Coulouris G, Zaretskaya I et al (2012) Primer-BLAST: a tool to design target-specific primers for polymerase chain reaction. BMC Bioinformatics 13:134. https://doi.org/10.1186/1471-2105-13-134
Altschul SF, Gish W, Miller W et al (1990) Basic local alignment search tool. J Mol Biol 215:403–410. https://doi.org/10.1016/S0022-2836(05)80360-2
Sievers F, Wilm A, Dineen D et al (2011) Fast, scalable generation of high‐quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol 7. https://doi.org/10.1038/msb.2011.75
Owen RW, Haubner R, Mier W et al (2003) Isolation, structure elucidation and antioxidant potential of the major phenolic and flavonoid compounds in brined olive drupes. Food Chem Toxicol 41:703–717. https://doi.org/10.1016/S0278-6915(03)00011-5
Muniandy K, Gothai S, Badran KMH et al (2018) Suppression of proinflammatory cytokines and mediators in LPS-Induced RAW 264.7 macrophages by stem extract of Alternanthera sessilis via the inhibition of the NF-κB pathway. J Immunol Res 2018. https://doi.org/10.1155/2018/3430684
Sabokbar N, Khodaiyan F (2016) Total phenolic content and antioxidant activities of pomegranate juice and whey based novel beverage fermented by kefir grains. J Food Sci Technol 53. https://doi.org/10.1007/s13197-015-2029-3
Bhat R, Suryanarayana LC, Chandrashekara KA et al (2015) Lactobacillus plantarum mediated fermentation of Psidium guajava L. fruit extract. J Biosci Bioeng 119. https://doi.org/10.1016/j.jbiosc.2014.09.007
Adebo OA, Gabriela Medina-Meza I (2020) Impact of fermentation on the phenolic compounds and antioxidant activity of whole cereal grains: a mini review. Molecules 25. https://doi.org/10.3390/molecules25040927
Li Z, Teng J, Lyu Y, et al (2018) Enhanced antioxidant activity for apple juice fermented with Lactobacillus plantarum ATCC14917. Molecules 24. https://doi.org/10.3390/molecules24010051
Zhou Y, Wang R, Zhang Y et al (2020) Biotransformation of phenolics and metabolites and the change in antioxidant activity in kiwifruit induced by Lactobacillus plantarum fermentation. J Sci Food Agric 100. https://doi.org/10.1002/jsfa.10272
Jimenez N, Curiel JA, Reveron I et al (2013) Uncovering the Lactobacillus plantarum WCFS1 gallate decarboxylase involved in tannin degradation. Appl Environ Microbiol 79. https://doi.org/10.1128/AEM.00840-13
Bahadori MB, Zengin G, Bahadori S et al (2018) Phenolic composition and functional properties of wild mint (Mentha longifolia var. calliantha (Stapf) Briq.). Int J Food Prop 21:183–193. https://doi.org/10.1080/10942912.2018.1440238
Zhao BT, Kim TI, Kim YH et al (2018) A comparative study of Mentha arvensis L. and Mentha haplocalyx Briq. by HPLC. Nat Prod Res 32:239–242. https://doi.org/10.1080/14786419.2017.1343325
Thumann TA, Pferschy-Wenzig E-M, Moissl-Eichinger C, Bauer R (2019) The role of gut microbiota for the activity of medicinal plants traditionally used in the European Union for gastrointestinal disorders. J Ethnopharmacol 245:112153. https://doi.org/10.1016/j.jep.2019.112153
Nakazawa T, Ohsawa K (1998) Metabolism of rosmarinic acid in rats. J Nat Prod 61:993–996. https://doi.org/10.1021/np980072s
Bel-Rhlid R, Crespy V, Pagé-Zoerkler N et al (2009) Hydrolysis of rosmarinic acid from rosemary extract with esterases and Lactobacillus johnsonii in vitro and in a gastrointestinal model. J Agric Food Chem 57:7700–7705. https://doi.org/10.1021/jf9014262
Mosele JI, Martín-Peláez S, Macià A et al (2014) Study of the catabolism of thyme phenols combining in vitro fermentation and human intervention. J Agric Food Chem 62:10954–10961. https://doi.org/10.1021/jf503748y
Baba S, Osakabe N, Natsume M et al (2005) Absorption, metabolism, degradation and urinary excretion of rosmarinic acid after intake of Perilla frutescens extract in humans. Eur J Nutr 44:1–9. https://doi.org/10.1007/s00394-004-0482-2
Lai KK, Lorca GL, Gonzalez CF (2009) Biochemical properties of two cinnamoyl esterases purified from a Lactobacillus johnsonii strain isolated from stool samples of diabetes-resistant rats. Appl Environ Microbiol 75:5018–5024. https://doi.org/10.1128/AEM.02837-08
Filannino P, Gobbetti M, De Angelis M, Di Cagno R (2014) Hydroxycinnamic acids used as external acceptors of electrons: an energetic advantage for strictly heterofermentative lactic acid bacteria. Appl Environ Microbiol 80:7574–7582. https://doi.org/10.1128/AEM.02413-14
Gaur G, Gänzle MG (2023) Conversion of (poly)phenolic compounds in food fermentations by lactic acid bacteria: novel insights into metabolic pathways and functional metabolites. Curr Res Food Sci 6:100448. https://doi.org/10.1016/j.crfs.2023.100448
Gonthier M-P, Remesy C, Scalbert A et al (2006) Microbial metabolism of caffeic acid and its esters chlorogenic and caftaric acids by human faecal microbiota in vitro. Biomed Pharmacother 60:536–540. https://doi.org/10.1016/j.biopha.2006.07.084
Peppercorn MA, Goldman P (1971) Caffeic acid metabolism by bacteria of the human gastrointestinal tract. J Bacteriol 108:996–1000. https://doi.org/10.1128/jb.108.3.996-1000.1971
Filannino P, Bai Y, Di Cagno R et al (2015) Metabolism of phenolic compounds by Lactobacillus spp. during fermentation of cherry juice and broccoli puree. Food Microbiol 46:272–279. https://doi.org/10.1016/j.fm.2014.08.018
Ricci A, Cirlini M, Calani L et al (2019) In vitro metabolism of elderberry juice polyphenols by lactic acid bacteria. Food Chem 276:692–699. https://doi.org/10.1016/j.foodchem.2018.10.046
Kimura Y, Okuda H, Okuda T et al (1987) Studies on the activities of tannins and related compounds, X. Effects of caffeetannins and related compounds on arachidonate metabolism in human polymorphonuclear leukocytes. J Nat Prod 50:392–399
Nakamura Y, Ohto Y, Murakami A, Ohigashi H (1998) superoxide scavenging activity of rosmarinic acid from Perilla frutescens Britton Var. acuta f. viridis. J Agric Food Chem 46:4545–4550. https://doi.org/10.1021/jf980557m
Poquet L, Clifford MN, Williamson G (2008) Effect of dihydrocaffeic acid on UV irradiation of human keratinocyte HaCaT cells. Arch Biochem Biophys 476:196–204. https://doi.org/10.1016/j.abb.2008.01.019
Gonthier M-P, Donovan JL, Manach C et al (2003) Microbial aromatic acid metabolites formed in the gut account for a major fraction of the polyphenols excreted in urine of rats fed red wine polyphenols. J Nutr 133:461–467. https://doi.org/10.1093/jn/133.2.461
Rios LY, Gonthier M-P, Rémésy C et al (2003) Chocolate intake increases urinary excretion of polyphenol-derived phenolic acids in healthy human subjects. Am J Clin Nutr 77:912–918. https://doi.org/10.1093/ajcn/77.4.912
Wittemer SM, Ploch M, Windeck T et al (2005) Bioavailability and pharmacokinetics of caffeoylquinic acids and flavonoids after oral administration of Artichoke leaf extracts in humans. Phytomedicine 12:28–38. https://doi.org/10.1016/j.phymed.2003.11.002
Jenner AM, Rafter J, Halliwell B (2005) Human fecal water content of phenolics: the extent of colonic exposure to aromatic compounds. Free Radic Biol Med 38:763–772. https://doi.org/10.1016/j.freeradbiomed.2004.11.020
Silva FAM, Borges F, Guimarães C et al (2000) Phenolic acids and derivatives: Studies on the relationship among structure, radical scavenging activity, and physicochemical parameters. J Agric Food Chem 48:2122–2126. https://doi.org/10.1021/jf9913110
Baeza G, Sarriá B, Mateos R, Bravo L (2016) Dihydrocaffeic acid, a major microbial metabolite of chlorogenic acids, shows similar protective effect than a yerba mate phenolic extract against oxidative stress in HepG2 cells. Food Res Int 87:25–33. https://doi.org/10.1016/j.foodres.2016.06.011
Huang J, de Paulis T, May JM (2004) Antioxidant effects of dihydrocaffeic acid in human EA.hy926 endothelial cells. J Nutr Biochem 15:722–729. https://doi.org/10.1016/j.jnutbio.2004.07.002
Wang J, Hodes GE, Zhang H et al (2018) Epigenetic modulation of inflammation and synaptic plasticity promotes resilience against stress in mice. Nat Commun 9:477. https://doi.org/10.1038/s41467-017-02794-5
Barthelmebs L, Divies C, Cavin J-F (2000) Knockout of the p -coumarate decarboxylase gene from Lactobacillus plantarum reveals the existence of two other inducible enzymatic activities involved in phenolic acid metabolism. Appl Environ Microbiol 66:3368–3375. https://doi.org/10.1128/AEM.66.8.3368-3375.2000
Carrasco JA, Lucena-Padrós H, Brenes M, Ruiz-Barba JL (2018) Expression of genes involved in metabolism of phenolic compounds by Lactobacillus pentosus and its relevance for table-olive fermentations. Food Microbiol 76:382–389. https://doi.org/10.1016/j.fm.2018.06.020
Pswarayi F, Qiao N, Gaur G, Gänzle M (2022) Antimicrobial plant secondary metabolites, MDR transporters and antimicrobial resistance in cereal-associated lactobacilli: Is there a connection? Food Microbiol 102:103917. https://doi.org/10.1016/j.fm.2021.103917
Acknowledgements
We thank Dr. Tomoko Amimoto, Natural Science Center for Basic Research and Development (N-BARD), Hiroshima University, for the measurement of GC–MS and NMR data.
Author information
Authors and Affiliations
Contributions
Conceptualization: Shrijana Shakya and Narandalai Danshiitsoodol; methodology: Shrijana Shakya and Narandalai Danshiitsoodol; formal analysis: Shrijana Shakya; writing—original draft preparation: Shrijana Shakya; writing—review and editing: Shrijana Shakya, Narandalai Danshiitsoodol, Masafumi Noda, and Masanori Sugiyama; supervision: Narandalai Danshiitsoodol. All authors have read and agreed to the published version of the manuscript.
Corresponding author
Ethics declarations
Ethics Approval
Not applicable.
Consent to Participate
Not applicable.
Conflict of Interest
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Shakya, S., Danshiitsoodol, N., Noda, M. et al. Role of Phenolic Acid Metabolism in Enhancing Bioactivity of Mentha Extract Fermented with Plant-Derived Lactobacillus plantarum SN13T. Probiotics & Antimicro. Prot. 16, 1052–1064 (2024). https://doi.org/10.1007/s12602-023-10103-4
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12602-023-10103-4