Abstract
The management of fungal diseases imposes an urgent need for the development of effective antifungal drugs. Among new drug candidates are the antimicrobial peptides, and especially their derivatives. Here, we investigated the molecular mechanism of action of three bioinspired peptides against the opportunistic yeasts Candida tropicalis and Candida albicans. We assessed morphological changes, mitochondrial functionality, chromatin condensation, ROS production, activation of metacaspases, and the occurrence of cell death. Our results indicated that the peptides induced sharply contrasting death kinetics, of 6 h for RR and 3 h for D-RR to C. tropicalis and 1 h for WR to C. albicans. Both peptide-treated yeasts exhibited increased ROS levels, mitochondrial hyperpolarization, cell size reduction, and chromatin condensation. RR and WR induced necrosis in C. tropicalis and C. albicans, but not D-RR in C. tropicalis. The antioxidant ascorbic acid reverted the toxic effect of RR and D-RR, but not WR, suggesting that instead of ROS there is a second signal triggered that leads to yeast death. Our data suggest that RR induced a regulated accidental cell death in C. tropicalis, D-RR induced a programmed cell death metacaspase-independent in C. tropicalis, while WR induced an accidental cell death in C. albicans. Our results were obtained with the LD100 and within the time that the peptides induce the yeast death. Within this temporal frame, our results allow us to gain clarity on the events triggered by the peptide-cell interaction and their temporal order, providing a better understanding of the death process induced by them.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Our global and industrialized world is facing disheartening problems in the area of human, animal, and plant health. In the medical area, diseases caused by the burgeoning antibiotic-resistant microorganisms menace us to return to the pre-antibiotic era, for example, thwarting medical advances in cancer and transplant treatments [1, 2]. This problem is aggravated by insufficient research and development of new antibiotics by big pharmaceutical companies as a result of the intrinsic process of producing and marketing a new antimicrobial such as research high costs, long development time, and low financial return due to sporadic use compared to chronic or diseases which require drugs of continuous use [3]. Another handicap for pharmaceutical companies is that the development of bacterial resistance can make a newly launched antibiotic obsolete in a short time, undermining the financial return of the pharmaceutical industry that developed it [1, 3]. This fact is clearly demonstrated by the dwindling approval of new antibiotics for clinical use by regulatory agencies in the last two decades, which contrasts strikingly with the increasing rate of resistant bacteria [4, 5].
Besides bacteria, fungi are also of growing concern to medical and environmental areas. In this last one, they have been reported as the cause of drastic fall in wild bat [6] and amphibian [7] populations with serious ecological consequences [8, 9]. In agriculture, the problem caused by fungi is also serious and can put food security at risk [10]. This scenario is very worrying because estimates for the next 20 to 30 years are of a significant increase in the world population, which should reach approximately 9.8 billion humans in 2050 [11]. This population will put more pressure on world food production, and if there is a failure in the production process there would be serious consequences for human health, food security and the economy of some countries whose gross domestic product is heavily based on agribusiness [11]. In the medical area, recent data estimate that fungal infections ranging in severity from superficial to often fatal systemic invasive infections, in addition to being neglected, have increased worldwide, and among them are fungal keratitis, cutaneous mycoses, and airway diseases caused by fungal spores [10, 12, 13]. Those studies estimate that a total of 300 million people suffer from a severe fungal infection each year and about 1.5 million die as a result of these infections worldwide [12, 13]. This scenario has another complicating factor, the number of elderly patients or patients undergoing medical interventions or living with HIV and Mycobacterium tuberculosis infections, as well as the number of immunocompromised patients due to cancer treatment or transplant recipients has been increasing rapidly and all these patients, because of their medical conditions, are more susceptible to fungal infections [12, 13]. Additionally, besides the increase in the number of cases of fungal infections, there is also the report of Candida [14, 15] and Aspergillus [15] species resistant to clinical antifungals, aggravating the scenario. Moreover, new species Candida auris multiresistant to clinical antifungals was reported in 2009 [16]. In a study with four countries, Pakistan, India, South Africa, and Venezuela, and in a total of 54 patients infected with C. auris, 93% of the 54 isolates were reported to be resistant to fluconazole, 35% to amphotericin B and 7% the echinocandins. Most worryingly, out of the 54 isolates, 41% were resistant to two classes of antifungals, and 4% were resistant to three classes [17]. This study also shows that C. auris is already spread across three continents. In addition to multiple resistance, this emerging species is persistent in the hospital environment as it resists disinfection protocols and is easily propagated among patients, making it a serious threat to human health [16].
Another great concern of the scientific community is the world’s post-COVID-19 pandemic future. SARS-COV2 co-infection with other pathogens, including fungi, has been reported to increase the difficulty of diagnosis, treatment, and prognosis of COVID-19, but also exacerbate disease symptoms and mortality [18,19,20]. Antibiotics overuse to treat COVID-19 patients contributed to the increase in antibiotic-resistant microorganisms [21,22,23,24].
The aforementioned data underline the urgent need to develop efficacious therapeutics and rational strategies for both the prevention and treatment of fungal diseases, especially opportunistic ones caused by the yeast of Candida genus. Amongst the given alternatives, antimicrobial peptides (AMPs) have been forecasted as possible new therapeutic substances [25, 26]. AMPs are short polymers of up to 100 L-amino acid residues, synthesized by ribosomes, arranged in a linear or cyclic configuration with vast variation in sequence, length, and structure, with a net positive charge at physiological pH, and amphipathic character. The antimicrobial epithet derives from their broad inhibitory activities on various microorganisms, including multi-drug resistant ones. They are constitutively produced or induced in response to the perception of a pathogen attack in complex organisms, where they are part of the innate immune response, and in microorganisms, they are produced to avoid or eliminate competitors [25]. We have used an AMP-based bioinspired peptide design strategy for therapeutic use [27,28,29]. In our previous study, we gleaned from correlation studies between the primary structure and biological activities of plant AMPs, namely defensins, the position, and the amino acids required for the biological activity of plant defensins and their derived peptides. Based on this molecular identification, we designed three peptides and tested their antimicrobial activity against opportunistic yeasts of the Candida genus, Saccharomyces cerevisiae, and tested their toxicity against murine macrophages and human monocytes [29]. We showed that the antifungal activity of three bioinspired peptides was improved by target alterations to increase both charge and hydrophobicity and, in general, exerted low toxicity on the tested mammalian cells [29]. We also started testing strategies to stabilize the bioinspired peptides and introduced D-enantiomer substitutions, which in comparison with the L counterpart, proved to be more potent, likely due to the greater resistance to protease degradation [29]. Our preliminary data also established relevant criteria for the clinical use of bioinspired peptides, such as the small size of biologically active peptides that minimizes the production costs and diminishes the risk of inappropriate host interactions, and increases stability for in vivo applications. In the present work, we further investigated the mechanism of action of three bioinspired peptides against opportunistic yeasts to provide an understanding of the intracellular changes triggered by the peptides as a prerequisite to pave the way for clinical trials [30].
Material and methods
All reagents were acquired from Merck unless otherwise stated.
Peptides
The bioinspired peptides A36,42,44R37,38γ32–46VuDef (dubbed RR), D-A36,42,44R37,38γ32–46VuDef (dubbed D-RR), and A42,44R37,38W36,39γ32–46VuDef (dubbed WR) were designed and prepared as described in Toledo et al. [29]. The purity of the peptides was confirmed as ≥ 95% [29].
Yeast strains and antimicrobial assay
The cultivation of the yeasts Candida tropicalis (CE017) and Candida albicans (CE022), and the antimicrobial assay were done as described in Toledo et al. [29]. In brief, fresh yeast cultures were grown at 30 °C for 24 h and were used to obtain a colony that was resuspended in Sabouraud broth (5 g/L peptone from meat, 5 g/L peptone from casein, and 20 g/L D( +)glucose). The cell number of this stock cell suspension was determined by direct cell counting in a Neubauer chamber (Labor Optik) under an optical microscope (Axio Imager.A2, Zeiss). The antimicrobial assay was performed on a sterile 96-well microplate (polystyrene, U-shaped bottom, Nunc, Thermo Scientific) and consisted of 2,000 cells/mL of each yeast, the lethal dose of each peptide (27.5 µM RR and 23 µM D-RR for C. tropicalis, and 27.5 µM WR for C. albicans) filter-sterilized (0.22 μm, Millex-GV, Millipore), and 100 μL (final volume) of Sabouraud broth. Lethal dose or LD100 was defined as the lowest peptide concentration that caused 100% cell death of the assay cell population compared to control in the absence of peptide by colony forming units (CFU). Controls were done in the absence of peptides. Blanks were done with culture medium only.
Kinetic analysis of yeasts’ cell death induced by the designed peptides
To determine the kinetics of the yeast cell death an assay was carried out to determine the minimum period necessary for the designed peptides, at their LD100, to cause the loss of cell viability. This assay was done as described in Toledo et al. [29] or briefly in the subsection “Yeast strains and antimicrobial assay,” with the difference that the entire content of the wells was washed in Sabouraud broth and plated at every 3 h, from 0 to 21 h on plates containing Sabouraud agar (5 g/L peptone from meat, 5 g/L peptone from casein, 20 g/L D( +)glucose, and 17 g/L agar), according to Soares et al. [31]. By 0 h we mean the time needed to set up the experiment, wash the cells and plate them, which takes approximately 5 min. The peptides D-RR and WR were also tested at 1 and 2 h. After plating, CFU were determined at 24 h of incubation at 30 °C h for C. tropicalis and C. albicans. Cell death was defined as loss of cell division ability and loss of clonogenic capacity in culture medium in the absence of the stressor for 24 h of incubation [32]. The percentage of cell viability loss was calculated according to {[(CFU of test samples × 100)/CFU of control] − 100}.
Standardization of the number of cells for optical microscopy assays
To validate the antimicrobial activity of the designed peptides, we determined their effect on larger cell density at the LD100 and the time of death of each peptide. Cell viability assay was performed with the parameters determined in subsections “Yeast strains and antimicrobial assay” and “Kinetic analysis of yeasts’ cell death induced by the designed peptides,” with 40,000 cells/mL, as described in Soares et al. [31].
Analysis of endogenous reactive oxygen species production
The detection of reactive oxygen species (ROS) by yeasts after treatment with the bioinspired peptides was performed in antimicrobial assays in the presence of the antioxidant agents L-ascorbic acid (AA), N-acetyl-L-cysteine (NAC), or γ-L-glutamyl-L-cysteinyl-glycine (GSH). The antimicrobial assay was carried out as described in the subsection “Kinetic analysis of yeasts’ cell death induced by the designed peptides,” with the following modifications: the LD100 of the peptides, 70 mM of the antioxidant agent AA (1 M stock solution) added jointly with the peptides, 5 mM NAC (0.6 M stock solution) or 3 mM GSH (0.5 M) both added 30 min prior to addition of the peptides, all dissolved in ultrapure water, filter sterilized (Millex-GV 0.22 μm, Merck Millipore), and incubated at the previously determined time of death of each peptide. Controls also were made with the addition of the antioxidant agents and a positive control (control+) constituted of yeast cells incubated with 333 mM of acetic acid (AcA) for 1 h at 30 °C. AcA was used as a control+ in this study because it is a well-known inducer of regulated cell death in fungi with apoptosis-like features, and therefore a parameter for comparing the effect of the designed peptides. After this period, the cell viability assay was performed as described in the subsection “Kinetic analysis of yeasts’ cell death induced by the designed peptides.” For RR, samples were diluted fifty times (50x) before plating. Different concentrations of the three antioxidants were previously tested to determine the highest concentration that does not hinder growth without being toxic to the yeasts according to subsections “Yeast strains and antimicrobial assay and Kinetic analysis of yeasts’ cell death induced by the designed peptides.” The antioxidant AA was also incubated in different time intervals in relation to the addition of the peptides, pre- or post-incubated for RR and D-RR, and pre-incubated for WR, within the conditions aforementioned.
The production of ROS was also evaluated by indirect fluorescence microscopy, using 2’,7’-Dichlorodihydrofluorescein diacetate (H2DCFDA, Calbiochem, EMD) according to Mello et al. [27]. Yeast cultures (40,000 cells/mL) were incubated with the designed peptides at their LD100 and at the beginning of the time of death, 1 h for RR and C. tropicalis, 30 min for D-RR and C. tropicalis, and 20 min for WR and C. albicans. Controls were done without the addition of the bioinspired peptides. A positive control (control+) constituted of yeast cells incubated with 333 mM of acetic acid (AcA) for 1 h at 30 °C was also performed to adjust the parameters of excitation intensity and exposure time for the acquisition of fluorescent images, and these parameters were used for all other treatments. For C. tropicalis a control was also done in the presence of 70 mM AA. After peptide treatment, control, and treated cells were incubated with H2DCFDA (20 μM) for 15 min at 30 °C, transferred to slides, and covered with coverslips. Visualization was performed by differential interference contrast (DIC) (Axio Imager.A2, Zeiss) equipped with fluorescence filters (450 – 490 nm for excitation and 500 nm for emission) and cell images were acquired using an AxioCam MR5 camera (Zeiss) and AxionVision LE software (version 4.8.2, Zeiss). The results were expressed as percentage of fluorescent cells calculated by cell counting, using the following criteria: 1st—counting of approximately 200 cells in random DIC and fluorescent fields that varied according to the incubation times of the assays; 2nd—if the number of cells was too low (a consequence of the treatment with the bioinspired peptides or with the positive control that causes the death of the yeast cells), 10 random fields for each treatment were counted. The percentage of fluorescent cells, an indication of oxidative stress, was calculated to ([average number of fluorescent cells × 100] /average number of cells observed in DIC) for each sample.
Morphological analysis of yeast cells
Cell morphology was visualized by optical microscopy, as described in the subsection “Standardization of the number of cells for optical microscopy assays” with some modifications. The incubation time for antimicrobial assay for C. tropicalis cells with the designed peptides at the LD100 was 1 and 6 h with RR, and 30 min and 3 h with D-RR, whereas C. albicans cells were incubated for 20 min and 1 h with WR. Yeast cells cultured in the medium without the addition of the peptides were considered controls. Furthermore, three positive controls were included: yeasts heated at 100 ºC for 1 min; yeasts incubated with Triton X-100 (2%, V/V) for 15 min; yeasts incubated with 333 mM AcA for 1 h. An assay with 70 mM of the antioxidant agent AA was used (see subsection “Analysis of endogenous reactive oxygen species production” for the description of the antioxidant). After the incubation period of each treatment, the cells were transferred to slides and, after 5 min, covered with coverslips and analyzed under an optical microscope.
Cell size measurements were performed by evaluating the length of the transversal and longitudinal axis of cells after the treatment described above using AxionVision LE software (tool Measure, Length). Fifty randomly selected cells for each treatment were used in different observation fields. To provide uniform measurements, the cells were selected in the same focal plane, and pseudohyphae, non-intact cells, and early budding cells were not considered.
Mitochondrial function assays
Mitochondrial functionality was evaluated by optical fluorescence microscopy using fluorescent probes MitoTracker Red FM (Thermo Fisher) and Rhodamine 123 (Sigma-Aldrich). The assay was carried out as described in the subsection “Analysis of endogenous reactive oxygen species production,” with modifications. Yeast cells were incubated with the designed peptides at the beginning and end of the time of death, 1 and 6 h for RR and C. tropicalis, 30 min and 3 h for D-RR and C. tropicalis, and 20 min and 1 h for WR and C. albicans. Positive control cells were incubated with 2% Triton X-100 for 15 min. For C. tropicalis, a control was also done in the presence of 70 mM of AA. Control and peptide-treated cells were incubated with 0.1 μg/mL of the fluorescent probes Mitotracker Red FM or Rhodamine 123 for 15 min at 30 ºC and observed under an optical microscope (subsection “Analysis of endogenous reactive oxygen species production”) equipped with a 581 nm for excitation filter and a 644 nm emission filter for Mitotracker Red FM and 506 nm excitation filter and a 530 nm emission filter for Rhodamine 123.
Cell proliferation reagent WST-1 (Roche) was used to access the mitochondrial activity of the yeast cells treated with the designed peptides [33]. The assay for each peptide was prepared according to subsection “Yeast strains and antimicrobial assay” with 80,000 cells/mL. Then, 10 μL of WST-1 reagent and 2 μL of 2,3,5,6-tetramethyl-1,4-benzoquinone (duroquinone) (final concentration 0.24 mM of a stock of 12.24 mM in dimethyl sulfoxide (DMSO)) as an electron couple reagent [34], were added to each sample. A blank was prepared with Sabouraud broth, WST-1, and duroquinone. The optical density was monitored at 450 nm for 20 min and 1 h for WR, 30 min, 2 h, and 3 h for D-RR, and 1, 3, and 6 h for RR in an ELISA microplate reader (Epoch, Biotek) running Gen5 software. The percentage of mitochondrial activity was calculated according to [(ABS450nm of test samples × 100)/ABS450nm of control]. Standardization of the minimum number of cells in the assay required for the detection of WST-1 salt conversion by spectrophotometer was previously made. To validate the antimicrobial activity of the designed peptides, we tested their toxic effect on larger cell density at the LD100 and time of death of each peptide as described in the subsection “Standardization of the number of cells for optical microscopy assays.”
Regulated cell death analysis
Metacaspase activity was analyzed by optical fluorescence microscopy using fluorescent probe FITC-VAD-FMK (CaspACE, FITC-VAD-FMK In Situ Marker, Promega). The assay was carried out as described in the subsection “Analysis of endogenous reactive oxygen species production,” with modifications. Yeasts were incubated with the designed peptides for 20 min, 1 and 6 h (depending on the yeast and peptide tested), and in the positive control, the cells were incubated with 333 mM AcA for 1 h. For C. tropicalis, a control was also done in the presence of 70 mM AA. Control and peptide-treated cells were incubated with 50 μM FITC-VAD-FMK for 15 min 30 °C, washed once in PBS, and observed under a fluorescence microscope (subsection “Analysis of endogenous reactive oxygen species production”) with a 490 nm for excitation filter and a 525 nm emission filter.
To identify whether the death pathway activated by the designed peptides in yeasts is a type of regulated cell death with apoptosis-like features, an antimicrobial assay was performed in the presence of the pan-caspase inhibitor Z-VAD-FMK (Promega). The antimicrobial assay was carried out as described in subsection “Yeasts and antimicrobial assay,” with the following modifications: the LD100 of the peptides, 50 μM Z-VAD-FMK (2 mM stock solution in DMSO) were used and incubated at the previously determined death times. Controls were also made with the addition of 50 µM Z-VAD-FMK. Cell viability assay was performed as described in subsection “Kinetic analysis of yeasts’ cell death induced by the designed peptides,” with the following modifications: cells were plated at the time of death for each peptide.
Chromatin condensation was examined by 1 µM DAPI staining (1 mg/mL stock solution in ultrapure water) in cells treated as described in the subsection “Analysis of endogenous reactive oxygen species production” with the following modifications: after the incubation times, the control cells, only cells and medium, and the peptide treated cells were previously permeabilized with 2% Triton X-100 for 2 min prior to DAPI staining for 15 min in the dark at 30 °C and observed under a fluorescence microscope.
Accidental cell death analysis
Accidental cell death (terminology proposed by Carmona-Gutierrez et al. [67] instead of necrosis) was analyzed by fluorescence microscopy using propidium iodide (PI) fluorescent dye. The assay and analysis of the results were carried out as described in subsection “Analysis of endogenous reactive oxygen species production,” with the following modifications: the yeasts were incubated with the peptides at their LD100 for 1 and 6 h for RR, 30 min and 3 h for D-RR, and 20 min and 1 h for WR. For positive control, cells were heated for 1 min at 100 °C. Controls and treated cells were incubated with 0.25 μg/mL PI for 15 min in the dark and observed under a fluorescence microscope with a 506 nm excitation filter and a 530 nm emission filter. This analysis was also done with time intervals of 10 and 15 min.
Statistical analysis
All experiments were carried out three times in triplicate, with the exception of optical microscopy assays, carried out in singlicate. A one-way ANOVA test was performed using GraphPad Prism version 8.0.2 for Windows. P < 0.05 is considered statistically significant. GraphPad Prism version 8.0.2 and PowerPoint (Microsoft Office 365) were used to illustrate the data.
Results
We first determined the time required for the bioinspired peptides to induce the death of the entire yeast cell population in the antimicrobial assay, i.e. 2,000 cells/mL at their respective LD100. C. tropicalis incubated with 27.5 μM RR caused a 99% decrease in colony forming units (CFU) within 3 h, and induced the death of the entire yeast cell population from 6 h onward, accordingly cell death time of RR was 6 h (Table 1). However, 23 μM D-RR induced the death of C. tropicalis cell population already from 3 h onward. This shorter time of death prompted us to verify D-RR antifungal activity at the time intervals of 1 and 2 h, revealing a 98% decrease in CFU at 1 h and a 99% reduction at 2 h; since shorter treatment times were not effective in killing all cell population, D-RR induced time of death was estimated as 3 h (Table 1). Surprisingly, 27.5 μM WR caused 97% decrease in CFU at 0 h, and the death of 100% of C. albicans cell population after 3 h (Table 1). We also tested the antifungal activity of WR at 1 and 2 h and found that WR caused 100% death of C. albicans cell population after 1 h thus establishing WR time of death of 1 h. By 0 h we mean the time needed to set up the experiment, wash the cells and plate them, which takes approximately 5 min. Based on these experiments we choose the time points that represent the beginning and the end of the time of death interval required for each peptide to induce the death of the assay cell population (based on Table 1) at their respective LD100. Therefore, the following assays were done with the LD100 and time of death parameters as follows: 27.5 μM RR/1 and 6 h for C. tropicalis, 23 μM D-RR/30 min and 3 h for C. tropicalis, and 27.5 μM WR/20 min and 1 h for C. albicans.
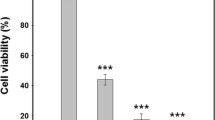
Before we started our study to unravel the mechanism of action of the bioinspired peptides on yeasts, we standardized the number of cells to validate our forthcoming fluorescence microscopy assays. Thus, an antimicrobial assay using 40,000 cells/mL, instead of 2000 cells/mL of the previous assays, was done to determine the toxicity of the bioinspired peptides at higher cell density. It is noteworthy that RR, D-RR, and WR at their same LD100 and time of death determined for the assay with 2000 cells/mL retained their toxicity to twenty times more cells than the original antimicrobial assay, causing cell viability loss in almost all cell population (Fig. S1). The CFU were not determined in control samples because of the excessive number of colonies formed, thus the cell death percentage was not calculated. With these results, we ensured that the signals observed at the microscopic analysis are signals that were triggered by the designed peptides that lead to yeast cell death.
Then, we proceeded to test the concentration of the antioxidants AA, NAC, or GSH to select the highest concentration that showed the lowest toxicity (inhibition of CFU in regard to the control) in a viability assay. We chose 70 mM of AA which for C. tropicalis incubated at the time of death of RR and D-RR had toxicity of 63% and 45.4% (Table 2), respectively, and the same concentration for C. albicans incubated during the time of death of WR did not present toxicity (Table 2). For NAC, we chose 5 mM which for C. tropicalis incubated with the time of death of RR and D-RR had a toxicity of 65.6% and 19.2% (Table 2), respectively, and the same concentration for C. albicans incubated at the time of death of WR had a toxicity of 3.6% (Table 2). For GSH, we chose 3 mM which for C. tropicalis incubated at the time of death of RR and D-RR had a toxicity of 33.3 and 34.8% (Table 2), respectively, and the same concentration for C. albicans incubated at the time of death of WR had a toxicity of 15.4% (Table 2). The viability assay in the presence of AA and C. tropicalis cells concurrently incubated with RR showed 62.4% of protection (Table 2), and concurrently incubated with D-RR the protection was 63.7% (Table 2) compared to the peptides alone (Table 1). For C. albicans cells concurrently incubated with AA and WR there was no protection (Tables 1 and 2). Two other antioxidants tested, namely NAC and GSH, were both pre-incubated 30 min before the addition of the bioinspired peptides, and presented a very low degree of protection to C. tropicalis from the toxic action of RR and D-RR (Table 2). Neither NAC nor GSH protected C. albicans cells from the toxic action of WR as they were not protected by AA (Table 2). The protection conferred by AA suggested that C. tropicalis was undergoing oxidative stress after treatment with RR and D-RR. The production of ROS was confirmed by the fluorescent probe H2DCFDA. C. tropicalis cells incubated with RR for 1 h presented 88.2% H2DCFDA positive cells, indicating that those cells were under oxidative stress induced by RR (Table 3 and Fig. S2a). C. tropicalis cells incubated with D-RR for 30 min presented 72.3% H2DCFDA positive cells, indicating that C. tropicalis cells were also under oxidative stress induced by D-RR (Table 3 and Fig. S2b). We also observed that H2DCFDA fluorescence signal in C. tropicalis cells treated with RR and D-RR was more intense and with a diffuse distribution pattern within the cell which was different from the AcA (control+) (Fig. S2a and 2b). C. albicans cells incubated with WR for 20 min presented 94.4% of fluorescent cells, indicating that those cells were also under oxidative stress induced by WR and the intensity of the fluorescence in regard to the AcA (control+) was weaker (Table 3 and Fig. S2c). Despite being under oxidative stress, C. albicans was not protected by any antioxidant tested. Because AA was the only antioxidant protecting C. tropicalis from death, we tested it concurrently incubated with C. tropicalis cells and RR in the microscopic analysis. This result showed a decline from 88.2 to 21.7% of cells with H2DCFDA positive signal, whose value was not different from control, therefore indicating a protection in comparison with cells treated only with RR (Table 3 and Fig. S2a). C. tropicalis concurrently incubated with D-RR and AA showed a dramatic drop from 72.3 to 1.1% of the cells under oxidative stress, indicating protection in comparison with cells treated with D-RR alone (Table 3 and Fig. S2b). Because all antioxidants tested did not protect C. albicans from the toxic effect of WR, the analysis of WR in the presence of an antioxidant was not done.
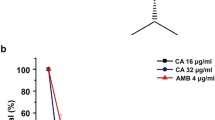
Because the antioxidant AA failed to protect the entire population of C. tropicalis cells from the toxicity of RR and D-RR in the viability test, we investigated why a fraction of the population of cells, even in the presence of the antioxidant, continued to die. For that, we tested the addition of AA in relation to the incubation time of the peptides, pre and post, in addition to the condition that had already been made of co-incubation. For WR and C. albicans, we only tested the pre-addition condition. AA added 30 min before the addition of RR to C. tropicalis, the percentage of cells not protected from death increased from 37.6 to 52.3% (Fig. 1A), to D-RR and C. tropicalis the percentage of cells not protected from death decreased from 36.3 to 10.6% (Fig. 1A). AA did not protect C. albicans cells from WR-induced death (Fig. 1C). AA added 1 h after the addition of RR and D-RR to C. tropicalis increased the percentage of cells not protected from death from 37.6 to 82.2% and from 36.3 to 68.6%, respectively (Fig. 1A and B). The percentage of C. tropicalis cells not protected from RR and D-RR death decreased even more 2 h after AA addition, reaching 98.4 and 98.9%, respectively (Fig. 1A and B).
Effect of the antioxidant ascorbic acid (AA) on the antimicrobial activity of the bioinspired peptides. AA was added before (pre), co-incubated (co), and 1 and 2 h after (post) the addition of RR (A) and D-RR (B) to Candida tropicalis, and before (pre) and co-incubated (co) the addition of WR (C) to Candida albicans cells. The colony forming units (CFU) values are means ± SD. The data from co-incubation were obtained from Table 1. **P < 0.01; *** P < 0.001; **** P < 0.0001. (ns) indicates not significantly different. The assay is representative of an independent assay out of three
The microscopic observations of cells treated with the designed peptides showed morphological alterations and thus they were further analyzed. We noticed that the cytoplasm of C. tropicalis, after 1 and 6 h of incubation with RR (Fig. 2A), and after 30 min and 3 h of incubation with D-RR (Fig. 2A), and C. albicans after 20 min and 1 h of incubation with WR (Fig. 2B), all had a granular and/or vacuolated appearance, whereas this morphological alteration was not detected in the control cells. This granularity of the cytoplasm was also observed in the positive control treatments, mainly those composed of AcA and heat (Fig. 2A and B). Although the antioxidant assays did not show protection for C. albicans (Table 2), we used AA as a control for these microscopic analyses. For the first 20 min after incubation, we did not observe changes in cell size, but cytoplasm granularity was attenuated, suggesting that ROS may be involved in the morphological alteration (Fig. 2B). For the time of death, 1 h, cytoplasm granularity, and size reduction were observed (Fig. 2B), which are in accordance with the non-protection of this yeast species by this antioxidant (Table 2). To confirm the overt cell shrinkage (Fig. 2A and B), we measured the longitudinal and transversal axes of the cells at their time of death. C. tropicalis cells treated with RR and D-RR presented a reduction of 29.7 and 23.2%, and of 34.9 and 27.4% for the longitudinal and transversal axes, respectively (Fig. 2C). C. albicans cells treated with WR presented a reduction of 38.3 and 33.8% for the longitudinal and transversal axes, respectively (Fig. 2D). AA protected C. tropicalis from the toxic effect of RR and D-RR (Table 2), and for RR concomitantly incubated with AA, it prevented 13.2% the longitudinal axis reduction, from 29.7 to 16.5%, and 18.9% the transversal axis reduction, from 23.2 to 4.3% (Fig. 2C). For D-RR concomitantly incubated with AA it protected 12.6% the longitudinal axis reduction, from 34.9 to 22.3%, and 23.6% the longitudinal axis reduction, from 27.4 to 3.8% (Fig. 2C). Although AA failed to protect C. albicans from death, we used it in this assay, and as observed before, the reduction of cell size was not inhibited and the size of the longitudinal and transversal axes were 38.4 and 49.5%, respectively, which were not significantly different from the peptide alone (Fig. 2D). AA protected C. tropicalis treated with RR and D-RR against cell shrinkage in comparison with the peptides alone (Fig. 2C). Although this protection in cell shrinkage was statistically different compared to control (cells and medium only) for the longitudinal axis (Fig. 2C) that indicates that the protection was partial. This partial protection was also observed in Table 2. The addition of AcA, an inducer of regulated cell death in fungi with apoptosis-like features, resulted in a reduction in the longitudinal and transversal axes by 19.5 and 9.5% for C. tropicalis, and 30.6 and 26% for C. albicans, respectively (Fig. 2C and D).
Morphological changes and size reduction in the yeast cells treated with the bioinspired peptides. A Microscopic images of Candida tropicalis cells treated with 27.5 µM RR and 23 µM D-RR. Scale bar represents 10 µm. AA, ascorbic acid, AcA, acetic acid. B Length of the longitudinal and transverse axes in C. tropicalis cells. C Microscopic images of Candida albicans cells treated with 27.5 µM WR. D Length of the longitudinal and transverse axes of C. albicans cells. A, B Note the granularity of the cytoplasm and the reduction in cell size. C, D The percentage numbers indicate a reduction of the axis in relation to their respective controls. Controls correspond to yeast cells cultivated in medium, control+ correspond to AcA or heat or Triton X-100 treatments. **** P < 0.0001. (ns) indicates not significantly different. The average size of each sample for C. tropicalis for longitudinal axis is: control 7.8 µm, AcA 6.3 µm, RR 5.5 µm, RR + AA 6.5 µm, D-RR 5.1 µm, D-RR + AA 6.1 µm. The average size of each sample for C. tropicalis for transversal axis is: control 4.7 µm, AcA 4.3 µm, RR 3.6 µm, RR + AA 4.5 µm, D-RR 3.4 µm, D-RR + AA 4.5 µm. The average size of each sample for C. albicans for longitudinal axis is: control 5.4 µm, AcA 3.7 µm, WR 3.3 µm, WR + AA 3.3 µm. The average size of each sample for C. albicans for the transversal axis is: control 3.9 µm, AcA 2.9 µm, WR 2.6 µm, WR + AA 2.0 µm. The assay is representative of an independent assay out of three
Next, we investigated mitochondrial functionality because ROS stemmed mainly from mitochondrial activity. C. tropicalis treated for 1 h with RR in the presence of Mitotracker Red FM showed that the cells had active mitochondrial membrane potential (Fig. 3A). However, a stronger and more diffuse fluorescent labeling of Mitotracker Red FM was observed in the treated cells when compared to the control or those treated with AA (Fig. 3A). The same result was observed for 6 h, i.e. the time of death induced by RR. However, the intensity of the fluorescent signal at this latter time was increased even more compared to the initial time point of 1 h (Fig. 3A). C. tropicalis treated for 30 min or 3 h with D-RR showed a slightly stronger fluorescent signal in the D-RR treated cells compared to the control (Fig. 3C). C. albicans treated for 20 min or 1 h with WR, we observed that likewise C. tropicalis treated with RR and D-RR, a slightly stronger fluorescent label of Mitotracker Red FM can be observed in the WR-treated cells compared to the control (Fig. 3E). The positive control with AcA also enhanced mitochondrial activity of both yeasts (Fig. 3B, D, and F), but to a less extent than the bioinspired peptides. To confirm the increase in the membrane potential, we performed WST-1 assay to analyze the mitochondrial metabolic activity. Our results showed that C. tropicalis treated with RR and D-RR had an increase of 305.2 and 565.1% in mitochondrial activity within the 1 h and 30 min of incubation, respectively (Fig. 3B and D). This higher activity was kept elevated for 3 and 2 h of incubation, respectively, and was still high at the time of death, 3 and 6 h of incubation, respectively (Fig. 3B and D). C. albicans treated with WR, an increase of 835.9% in mitochondrial activity was also observed within the 20 min of incubation, which was still high, 376.4%, at 1 h, the time of death (Fig. 3F). Triton X-100, which impaired mitochondrial accumulation of the Mitotracker Red FM probe (Fig. 3A, C, and E), also completely blocked WST-1 reduction in the tested yeasts (Fig. 3B, D, and F); however, the AcA and Triton X-100 treatments were not statistically different from their respective controls.
Mitochondrial functionality in opportunistic yeast (A, B, C, and D Candida tropicalis, E and F Candida albicans) cells treated with bioinspired peptides RR, D-RR, and WR lethal dose. A, C, and E Microscopic images of yeast cells; Mitotracker Red FM fluorescence in the cytoplasm indicates mitochondrial functionality. Scale bar represents 10 µm. Note the hyperpolarization of the mitochondrial membranes of the treated yeast cells. Ascorbic acid (AA) reversed the hyperpolarization. Control+ corresponds to Triton X-100 treatment. Controls correspond to yeast cells cultivated in a medium. B, D, and F Determination of mitochondrial metabolic activity using WST-1. The values above the test bars indicate the mitochondrial activity compared to the control. Controls correspond to yeast cells cultivated in medium, control+ correspond to acetic acid (AcA) or Triton X-100 treatments. **** P < 0.0001; The assay is representative of an independent assay out of three
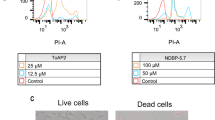
We also evaluated whether there was activation of metacaspases in the yeasts treated with the peptides with CaspACE FITC-VAD-FMK. For C. tropicalis treated with RR and for C. albicans treated with WR no positive signal was observed at the initial time of induction of death by these peptides (Figs. 4A, B, and 5A, B). C. tropicalis cells incubated with D-RR for 30 min presented 94.2% of FITC-VAD-FMK positive cells, indicating that C. tropicalis cells had activated metacaspases (Fig. 6A and B). For D-RR and C. tropicalis, the induction of metacaspase was inhibited by the coincubation of the cells with AA (Fig. 6A and B). The positive control (AcA) induced metacaspase activation in both yeast species (Figs. 4A, 5A, and 6A). The contribution of metacaspases activation in the death induced by the bioinspired peptides was examined by the pan-caspase inhibitor Z-VAD-FMK in the antifungal assay. It did not protect the tested yeasts from the action of the bioinspired peptides (Figs. 4C, 5C, and 6C). The analysis of chromatin condensation revealed that control cells presented a round shaped nucleus with an even DAPI fluorescent signal, whereas the peptide-treated cells exhibited a stronger and more punctual staining (Figs. 4D, 5D, and 6D). Fluorescent staining was even more conspicuous at the latter incubation times (Figs. 4D, 5D, and 6D). The same fluorescent staining pattern was also observed for AcA, a known inducer of regulated cell death in yeast with apoptosis-like features (Figs. 4D, 5D, and 6D). To better differentiate the type of cell death induced by the bioinspired peptides we investigated whether the death was an accidental cell death (ACD) by propidium iodide (PI) labeling. Our results indicated that C. tropicalis treated with RR and C. albicans treated with WR both showed positive labeling for PI since the beginning of incubation and also at the ending of the time of death (Table 4, and Fig. S3A and C) reflecting the presence of necrotic cells. Conversely, C. tropicalis cells treated with D-RR were PI negative (Table 4 and Fig. S3B). Additionally, for C. tropicalis incubated with RR, the PI positive labeling was reversed by AA co-treatment (Table 4 and Fig. S3A). Noteworthy, the presence of PI positive cells pointed to another result of peptide effect: the plasma membrane permeabilization in C. tropicalis treated with RR and in C. albicans treated with WR, but not for D-RR-exposed C. tropicalis cells. As the labeling of PI would help us to better differentiate the effect of peptides, we tested early time points for RR and WR and found that PI labeling starts for both peptides and their respective yeasts after 15 min of incubation (Table 5).
Cell death analysis in Candida tropicalis cells treated with RR lethal dose. A Microscopic images of yeast cells; FITC-VAD-FMK green fluorescence in the cytoplasm indicates metacaspase activation. Scale bar represents 10 µm. B Number of cells with activated metacaspases, determined after cell count in random DIC and fluorescence fields as observed in A. Different letters indicate a statistically significant difference. P < 0.05. C Viability assay with C. tropicalis incubated with RR at its time of death (6 h) and control in the presence of the pan-caspase inhibitor Z-VAD-FMK. Colony forming units (CFU) values are means ± SD. (nd), not determined (excessive number of grown colonies that prevented colony counting). (D) Detection of chromatin condensation by DAPI staining. Note that the control cells present a round-shaped nucleus with an even fluorescent signal, whereas the peptide-treated cells present a stronger and more punctual staining indicating chromatin condensation. Scale bar represents 10 µm. A–D Controls correspond to yeast cells cultivated in medium. A, B, D control+ corresponds to acetic acid (AcA) treatment. The assay is representative of an independent assay out of three
Cell death analysis in Candida albicans cells treated with WR lethal dose. A Microscopic images of yeast cells; FITC-VAD-FMK green fluorescence in the cytoplasm indicates metacaspase activation. Scale bar represents 10 µm. B Number of cells with activated metacaspases, determined after cell count in random DIC and fluorescence fields as observed in A. Different letters indicate a statistically significant difference. P < 0.05. C Viability assay with C. albicans incubated with WR at its time of death (1 h) and control in the presence of the pan caspase inhibitor Z-VAD-FMK. Colony forming units (CFU) values are means ± SD. (nd), not determined (excessive number of grown colonies that prevented colony counting). D Detection of chromatin condensation by DAPI staining. Note that the control cells present a round shaped nucleus with an even fluorescent signal, whereas the peptide-treated cells present a stronger and more punctual staining indicating chromatin condensation. Scale bar represents 10 µm. A–D Controls correspond to yeast cells cultivated in medium. A, B, D control+ corresponds to acetic acid (AcA) treatment. The assay is representative of an independent assay out of three
Cell death analysis in Candida tropicalis cells treated with D-RR lethal dose. A Microscopic images of yeast cells; FITC-VAD-FMK green fluorescence in the cytoplasm indicates metacaspase activation. Scale bar represents 10 µm. B Number of cells with activated metacaspases, determined after cell count in random DIC and fluorescence fields as observed in A. Different letters indicate a statistically significant difference. P < 0.05. A, B Ascorbic acid (AA) reversed the metacaspase activation. C Viability assay with C. tropicalis incubated with D-RR at its time of death (3 h) and control in the presence of the pan caspase inhibitor Z-VAD-FMK. Colony forming units (CFU) values are means ± SD. (nd), not determined (excessive number of grown colonies that prevented colony counting). D Detection of chromatin condensation by DAPI staining. Note that the control cells present a round-shaped nucleus with an even fluorescent signal, whereas the peptide-treated cells present a stronger and more punctual staining indicating chromatin condensation. Scale bar represents 10 µm. A–D Controls correspond to yeast cells cultivated in medium. A, B, D control+ corresponds to acetic acid (AcA) treatment. The assay is representative of an independent assay out of three
Discussion
The broad spectra of microbial inhibitions of AMPs catapulted them to a new pharmaceutical status. Nevertheless, the lack of understanding of their in vivo performance imposes some weaknesses on their pharmaceutical development and stalling their way to the clinic. Such drawbacks are embodied by their toxic action against mammalian cells, which results in a low selective index, poor in vivo pharmacokinetic and pharmacodynamic that render low bioavailability to AMPs, and high production costs, due to their complex structure and length [35]. But embedded in their structures are features that are being explored to circumvent these drawbacks such as wide structural diversity that coupled with correlation studies between activity and structure are showing both an understanding of positions and nature of amino acids important for activity, as well as, the possibility by chemical synthesis of incorporating unnatural amino acids, improving their stability and activity [36]. These understandings allowed the design of modified AMPs with better therapeutic properties. We used a similar approach correlating the primary structure and biological activities of plant defensins which allowed us to design three new peptides, named RR, D-RR, and WR, with ameliorated antimicrobial activity against opportunistic yeasts and low toxicity toward mammalian cells [29]. Herein, we further characterize these bioinspired peptides and set in motion a series of experiments to unravel the possible mechanism of their action against the opportunistic yeasts in regard to the induced events which led to cell death.
Previously we determined that LD100, which causes 100% of yeast death, was 27.5 μM and 23 μM of RR and D-RR, respectively, for C. tropicalis, and 27.5 μM WR for C. albicans [29]. Herein we determined the time of death (Table 1) and then chose one time in the beginning and another at the end of the time of death for the next experiments. These parameters are important because they avert misinterpretation of the cell death pathway induced by the bioinspired peptides [31]. Afterwards, we standardized the number of cells for microscopy analysis. The antimicrobial assays carried out so far used a total of 2000 cells/mL; however, this amount of cells is unfeasible for optical microscopy analysis because it is below the detection limit. Therefore, a higher amount of cells was used to allow their visualization under an optical microscope. For that reason, 40,000 cells/mL were used based on Soares et al. [31]. The bioinspired peptides retained their toxicity at higher yeast cell numbers (Fig. S1).
Allegedly, the main mechanism of action involved in microbial growth inhibition or death by AMPs is microbial cell membrane damage. Indeed some AMPs have been reported to interact with microbial membrane components such as phospholipids and sphingolipids [37,38,39]. Contradictory, some works reported results that do not fully explain that the AMP-membrane interaction is the main event responsible for the microbial growth inhibition or cause of death, and others demonstrated that the membrane damage is a ripple effect, i.e. the microbial membrane is a secondary or indirect target of AMPs action with other primary targets [31, 40,41,42,43]. Moreover, other mechanisms of action have been described [44]. However, the interaction of AMPs with microbial membrane components seems to be crucial for growth inhibition and/or death processes. Amidst them are the anchorage points for AMPs at microbial membrane components by opposite charge attraction which allow their accumulation on the membrane surface and their latter entrance into the cell. This view found support in the reports that some known mechanisms of bacterial resistance to AMPs are mediated by camouflaging negative charges of both membrane lipid and cell wall components [45, 46]. One such mechanism of action of several antimicrobial substances, including AMPs, to which the inhibitory activity or the induced death upon microorganisms seems to converge, is the induction of endogenous oxidative stress [47, 48].
Firstly, based on those reports of oxidative stress as the possible primary toxic effect, we analyzed whether bioinspired peptides promoted endogenous oxidative stress in the yeast cells. For that, we analyzed the contribution of oxidative stress to the death induced by the designed peptides by treating the yeasts with the antioxidant agents AA, NAC, or GSH in a viability assay. These substances are non-enzymatic antioxidants capable of protecting cells against the oxidizing action of free radicals [49, 50]. Thus, this analysis makes it possible to infer whether there is ROS generation in yeast cells exposed to the bioinspired peptides and, additionally, to elucidate their mechanistic involvement in yeast death. Our results indicated protection from death to C. tropicalis incubated with RR and D-RR, and no protection for C. albicans incubated with WR (Table 2). Antioxidant AA were shown to diminish the antifungal activities of NaD1 (plant defensin from Nicotiana alata) [51], Rs-AFP2 (plant defensin from Raphanus sativus) [41], and ApDef1 (plant defensin from Adenanthera pavonina) [31] against C. albicans and S. cerevisiae. That family of plant AMP is which RR, D-RR, and WR were bioinspired [29], and thus it is expected they share the mechanism of action. Those reports not only are in accordance with our results but also indicate that oxidative stress is involved in the cell death induced by these peptides. Antioxidant NAC is widely used as an ROS scavenger in yeast cells [50, 52, 53] and was shown to protect C. albicans from the toxic effect of itraconazole at 5 mM [50], to protect C. albicans and S. cerevisiae from the toxic effect of KM29 (synthetic AMP derived from Homo sapiens histatin 5) at 10 mM [53], and to protect C. albicans from the toxic effect of LfcinB15 (AMP derived from the proteolytic cleavage of lactoferrin) at 60 mM [39]. Nonetheless, it had no effect against the toxic effect of 10 mM farnesol in S. cerevisiae [54]. In these three examples, the antioxidant activity was associated with ROS toxicity, to the same extent inferred in our results of co-incubation of RR and D-RR with the antioxidant AA. Having said that, in our test conditions 3 mM NAC was unable to rescue yeast cells from induced death (Table 2). Additionally, in our test conditions, at concentrations higher than 5 mM NAC was toxic for C. tropicalis and C. albicans, completely abrogating the growth of yeast cells (data not shown). Indeed, Andrés et al. [55] reported the toxic effect of NAC on C. albicans at concentrations higher than 15 mM, and AA toxicity for Candida was also reported [56]. This toxicity action of the antioxidants on our conditions occurred most likely because the oxidative status of the cells has important signaling roles [57,58,59] and higher concentrations of exogenous antioxidants might preclude these signaling pathways. One possible explanation for the lower protection of C. tropicalis and no protection of C. albicans by NAC is that NAC is an optimal scavenger for hydroxyl anion (OH•) [60]. If it is the case, it points to an irrelevant participation of OH• in the peptide-induced death or non-production of it by the yeast species in the conditions studied. Another possibility for the antioxidant activity of NAC is an indirect effect as a precursor used by the cell to synthesize GSH, which ultimately has antioxidant action [61]. At all, GSH synthesis may not have occurred fast enough to confer antioxidant protection by increasing GSH content since the addition of the peptides, especially WR which toxicity was observed after 0 h of cell contact (Table 1), although GSH failed to rescue the tested yeasts from the toxic action of the bioinspired peptides. GSH by itself is a well-known antioxidant molecule in fungi [62, 63]. Its addition was reported to protect C. albicans from death induced by LfcinB15 [40], but in high concentrations, it was toxic to S. cerevisiae by extra-mitochondrial Fe–S enzymes maturation [64]. In our test conditions, GSH at 3 mM or higher concentrations negatively impacted C. tropicalis growth (33.3% and 34.8% for RR and D-RR, respectively, Table 2) and C. albicans (15.4%, Table 2), and therefore, we had chosen in our condition tested the highest GSH concentration that was minimally toxic for the tested yeast cells.
To confirm the endogenous induction of ROS by the yeasts, we used the fluorescent indicator H2DCFDA. This probe is cell-permeable and once inside the cell it is deacetylated by intracellular esterases and, when oxidized by ROS, generates the fluorescent compound dichlorofluorescein allowing the detection of ROS inside the cell. AcA was used as a positive control in this assay because it is a well-known inducer of regulated cell death in fungi with apoptosis-like features [65, 66]. In the conditions tested, AcA caused 100% of cell death (Table S1), which confirms its cell death induces capability [66]. Additionally, for this fluorescent analysis, cells treated with AcA were used as parameters to adjust the microscope for the control and peptides treatments and all images were captured with the exposure time and excitation intensity adjusted for this positive control (Fig. S2). The time points chosen represented one point at the beginning of the time of death interval required for each peptide to cause the death of the assay cell population (based on Table 1). Our results indicate that RR and D-RR, and WR induced oxidative stress in C. tropicalis and C. albicans, respectively (Table 3 and Fig. S2), and antioxidants use in the viability assay not only confirm our assumption that C. tropicalis cells were undergoing oxidative stress resulting from the action of RR and D-RR, but also that ROS is related to the signal triggered by RR and D-RR that led to cell death, provided that once scavenged by the antioxidant, it rescued C. tropicalis cells from death (Table 2). Moreover, C. tropicalis death was pharmacologically inhibited, suggesting the occurrence of a regulated cell death (Table 2) [67]. It is of note that the presence of the three different antioxidants did not rescue C. albicans from death, even despite cells facing oxidative stress. One explanation might be the rapid rate with which WR induced the death of this yeast species, since immediately after WR addition (0 h time of death), 97% of the cell population was already dead (Table 1). Such rapidity may prevent the antioxidant from having the protective effect even when added before WR (also see discussion of Fig. 1C). Another possibility to explain the absence of antioxidant protection effectiveness is discussed later in this section.
During the analysis of our results of the oxidative stress determination assays, we noted that 37.6 and 36.3% of C. tropicalis cells died even in the presence of the AA when incubated with RR and D-RR, respectively (Table 2). These results indicate the antioxidant rescues 62.4 and 63.7% of C. tropicalis cells from death (Table 2). The death of 37.6 and 36.3% of C. tropicalis cells even in the presence of the antioxidant AA (Table 2) raises the question of why the antioxidant did not protect 100% of the assay cell population. The same result was observed in the microscopic assay in which C. tropicalis treated with RR and D-RR, simultaneously with AA, did not present a complete reversal of cell size reduction (Fig. 2). Even considering that yeast cells do not have a synchronized division cycle [68], AA should have a higher protection rate if the oxidative stress was the main event leading to peptide-induced yeast death. Based on this incomplete protection, our assumption was that ROS might be an initial stress that once established triggers another cell death executor(s) that lead to the cell death. This assumption explains that even in the presence of the antioxidant, and therefore protected from ROS, yeast cells would still be committed to death. To discriminate this effect, we repeated the antimicrobial and viability assays in the presence of the antioxidant AA, but adding it at different times in relation to the addition of RR and D-RR. AA added 30 min before the addition of RR to C. tropicalis, which in theory should provide greater protection against ROS since the cell would be preloaded with the antioxidant and it would promptly eliminate the generated ROS, the percentage of cells not protected from death, unexpectedly, increased from 37.6 to 52.3% (Fig. 1A). We believe that this decrease in protection may be due to the intrinsic toxicity of AA, which in this case lasted a total of 6 h 30 min of incubation with the yeast C. tropicalis, 30 min longer than the co-incubation condition and, therefore, ended up being more toxic. In fact, comparing the control co-incubated with AA with the control pre-incubated with the antioxidant we observed a reduction of 17.2% indicating toxicity (Fig. 1A). To D-RR and C. tropicalis, as foreseen, the pre-incubation for 30 min with AA, the percentage of cells unprotected from death decreased from 36.3 to 10.6% (Fig. 1B). This higher toxicity observed to RR due to the longer incubation time is also evident when compared with the results of D-RR and the same yeast, in this case, the total incubation time was 3 h and 30 min. AA added 1 h after the addition of RR to C. tropicalis, i.e. ROS would have enough time to act on the cell, and the percentage of cells not protected from death, as expected, increased from 37.6 to 82.2% (Fig. 1A). In the case of D-RR, AA was added 1 h after the addition of the peptide and the percentage of cells not protected from death also increased from 36.3 to 68.6% (Fig. 1B), and added 2 h after the addition of the RR and D-RR, the percentage of cells not protected from death dropped even more, reaching 98.4 and 98.9%, respectively (Fig. 1A and B). For WR we adopted a different approach, as this peptide induced the death of C. albicans very rapidly (97% of cell death at 0 h of incubation, Table 1), we added AA only 30 min before the addition of the peptide because when AA was added jointly with WR it did not improve cell death (Table 2), and for this reason, there was no sense to add it later. However, even added 30 min before WR, AA continued to show the same response as when added concomitant to the peptide, no protection (Fig. 1C). C. albicans treatment with WR resulted in oxidative stress as confirmed by the fluorescent microscopy analysis (Fig. S2C) and since the peptide induced death of 97% of the cell population of the assay at 0 h (Table 1), we suspect that strong and rapid toxic effect of WR over C. albicans may enshroud the protection of the antioxidant even if it added before WR (Fig. 1C). For RR and D-RR, these results, as predicted, showed that ROS is probably not the final executor of C. tropicalis death induced by the designed peptides since once it was scavenged by the antioxidant after 1 and 2 h after the addition of the peptides, C. tropicalis was still committed to death. This result also raised the question of the nature of another cell death executor triggered instead of ROS or whether there would be a mechanism corresponding to a point-of-no-return during the death process. The point-of-no-return for C. tropicalis treated with RR seems to take place before 3 h because at this time 98% of cells were dead, for C. tropicalis treated with D-RR this point likely occurred before 1 h because at this time 98% of cells were dead, and for C. albicans treated with WR seems to be after 5 min, because at 0 h, our time point correspondent to 5 min, 97% cells were dead (Table 1). Future studies should address the point-of-no-return or the other cell death signal. On the other hand neither the antioxidants used protected C. albicans from WR toxic action, suggesting that oxidative stress is not the main mechanism that leads to cell death despite being involved in the process.
Cell morphological analysis indicated that the yeasts treated with the bioinspired peptides exhibited granular cytoplasm and cell shrinkage (Fig. 2). Noteworthy the same phenotypic feature, i.e. cytoplasm granularity, was also induced by AcA, well-known inducer of regulated cell death in fungi with apoptosis-like features [66], which, jointly with the results from oxidative stress assays (Table 1), reinforce that regulated cell death may be occurring in the peptide-treated yeasts. Another interesting feature of the treated cells was their shrinkage compared to the control cells (Fig. 2A and B). Both the cytoplasmic alterations and cell shrinkage were observed from the early time until the time of death of the tested yeasts (Fig. 2A and B). Furthermore, it is interesting to note that C. tropicalis cells treated with RR and D-RR along with the antioxidant AA did restore both cytoplasmic changes and apparent size reduction (Fig. 2A and B), reinforcing again that these toxic effects of RR and D-RR are mediated by ROS and also pointing out to ROS as the primary stress that leads to both the cytoplasmic changes and cell size reduction. Our measurements confirm the visually observed cell size reduction (Fig. 2C and D). As the same effect, i.e. cell shrinkage, was observed for both the bioinspired peptides and AcA, it is evidencing once again a type of regulated cell death induced by the designed peptides. Cell shrinkage is a well-known hallmark phenotype of several types of regulated cell death in mammalian cells [49]. In fungi, cell shrinkage was characterized during death induced in C. albicans by exogenous molecules like chlorogenic acid [69], and histatin 5 (AMP from Homo sapiens saliva) [70]. In both examples, the efflux of ions, especially K+, was pointed out as the cause of cell shrinkage. If this is the case, it raises two possibilities; first, K+ efflux is the cause of cell shrinkage, and second, it may be the other signal that is independent of the antioxidant action triggered by the bioinspired peptides, and both possibilities should be investigated in the future studies. Taken together, the death induction by the bioinspired peptides (Table 1), the induction of ROS (Table 2), and cell size reduction (Fig. 2) strongly suggest that designed peptides induced a type of regulated cell death with apoptosis-like features in the tested Candida yeasts.
The production of ROS identified in yeasts after treatment with the designed peptides led us to analyze their mitochondrial functionality because mitochondria are the main source of intracellular ROS as a byproduct of cellular respiration due to a leakage of electrons from the determined flow order on the electron transport chain (ETC) directly to molecular oxygen (O2) [71, 72]. Therefore, we used the membrane potential-dependent probes Mitotracker Red FM and Rhodamine 123 to assess mitochondrial functionality. All three peptides induced mitochondrial dysfunction in the Candida yeasts (Fig. 3). These results also indicate by the higher intense fluorescent signal that the alteration induced by the peptides already took place at the beginning of the induced cell death process, i.e. 1 h for RR, 30 min for D-RR and 20 min for WR (Fig. 3). It must be emphasized that the comparison of control and peptide-treated samples within the same time point can be interpreted relative to one another because the images were acquired with the same fluorescent intensity and exposure time settings, adjusted to their respective controls (cells and medium). The same results, i.e. the increase in the fluorescent intensity signal, were obtained for Rhodamine 123 for all peptides and treated yeasts (data not shown). These results point out that the mitochondria of the peptide-treated yeasts underwent dysfunctional stress and for C. tropicalis the anomalous strong signal of the probes, i.e. Mitotracker Red FM or Rhodamine 123, was directly linked to the ROS production since co-incubation of AA with RR or D-RR in C. tropicalis restored the intensity of the fluorescent signal equal to the control cells for both probes (Fig. 3 and Fig. S2). Additionally, AA also reduced the intensity of the fluorescent signal of ROS probe, that is, the antioxidant relieved the oxidative stress on the mitochondria (Fig. S2). The increased uptake of the mitochondrial probes observed for the peptide-treated yeasts in this work may indicate a hyperpolarization of the mitochondrial membrane potential. It is also in accordance with the intensified ROS probe signal which also showed an increase in fluorescence intensity as a result of intense ROS production (Fig. S2A and B). These results seem to be related because ROS production depends on a flow of electrons in the mitochondria ETC [73,74,75]. For a description of mitochondrial ETC please refer to Vendetti et al. [73], Mailloux [74], and Napolitano et al. [75]. One known condition that leads to ROS production in complex I is by reverse electron transport (RET). In RET the mitochondrial membrane must have a high pool of reduced coenzyme Q (CoQ) and a high mitochondrial membrane potential ∆ψm which act together to reverse the flow of electrons back to complex I where at the flavin mononucleotide site, electrons are passed to O2 generating superoxide anion [76]. C. albicans and C. parapsilosis are known to possess all five mitochondrial complexes which are denominated classical respiratory pathways, and in addition, two other known pathways were described in these yeasts species denominated as alternative and parallel oxidases [77]. Those last two are considered compensatory pathways when the classical pathway is somehow blocked [78]. The alternative and parallel pathways branch off from the classical pathway at the site of CoQ, therefore they can deoxidize CoQ, thus blocking one of the RET requirements from happening in these yeasts [76, 78]. Some works have demonstrated that the main site of ROS generation in fungi is the mitochondrial complex I [78,79,80]. Some reports have positively correlate high ∆ψm and low ATP concentration with an increase in endogenous ROS production to the toxic effect of antifungal substances in Candida species, for example, in a C. albicans fluconazole-resistant strain treated with a combination of fluconazole and berberine [76, 81], and C. albicans treated with plagiochin E [82]. The high ∆ψm has also been associated with ROS production in hyperpolarized mitochondria of S. cerevisiae treated with the isoprenoid farnesol [54]. Also, protamine sulfate, a drug used in cardiovascular surgery causes a reduction in cardiac oxygen consumption. Its mechanism of action is related to the inhibition of cytochrome c oxidase activity causing respiration inhibition, hyperpolarization of mitochondrial membrane potential, and ROS production [83]. The increase in mitochondrial membrane potential for ROS generation is corroborated by Skulachev [84] who highlighted that for ROS production in both complex I and complex III, an increase in mitochondrial membrane potential is required. Accordingly, our results also correlate with the increase in mitochondrial potential and increased ROS production (Figs. 3 and S2). But what increases the mitochondrial potential in the first place? A possible explanation was given by Pozniakovsky et al. [85], who correlated the increase in mitochondrial potential with the transient rise in intracellular Ca2+ in S. cerevisiae treated with the antiarrhythmic agent amiodarone, which in turn increased the activity of mitochondrial NADH dehydrogenases, which ultimately supply electrons to the ETC chain and consequently to ROS production. This explains both the increase in mitochondrial membrane potential and the increase in ROS production by the increase in the flow of electrons. The increased expression of isocitrate dehydrogenase, an enzyme of the Krebs cycle, in C. albicans treated with fluconazole and berberine corroborates with the observation of Pozniakovsky et al. [85, 80]. Another possibility for the hyperpolarization of mitochondrial membrane potential was given by Machida and Tanaka [54] who indicated that farnesol precludes cellular signaling which results in the spurring F1-ATPase while reversing its activity from ATP synthesis to ATP hydrolysis in S. cerevisiae, resulting in proton ions transport back into the intermembrane space, hyperpolarization of the mitochondrial membrane potential and ROS production. It is remarkable that this mitochondrial stress scenario with membrane hyperpolarization nor only tentatively explains the toxic effect of the designed peptides towards C. albicans and C. tropicalis, but also explains why S. cerevisiae that lacks the complex I of the classical respiratory pathway, together with alternative and parallel pathways [86] is resistant to the designed peptides [29], provided that complex I is the site of ROS generation. To confirm the high mitochondrial activity, already observed with the Mitotracker Red FM (Fig. 3A, C, and D) and Rhodamine 123 probes (data not shown), we used WST-1 assay to assess mitochondrial activity on the peptide-treated yeasts based on studies showing that the NADH formed by the mitochondrial Krebs cycle is the leading reductant for WST-1 reduction to water-soluble formazan outside cells in the presence of an intermediate electron acceptor, allowing the quantification of mitochondrial metabolic activity by colorimetric assay [33]. The high activity observed in the reduction of WST-1 at the yeasts’ time of death may represent a residual activity of the enzymes involved in the process [66]. Therefore, the results of the WST-1 assay support the results of the stronger staining of the membrane potential-dependent fluorescent probes and confirm that the mitochondria of treated yeasts were hyperpolarized. For the WST-1 assay, we had to standardize the minimum number of cells to give a positive signal in the assay. Initially, we used the 40,000 cells/mL that was already normalized for the microscopy assays (Fig. S1) validating that the peptides had activity at higher cell concentration, but we obtained no positive results within the death incubation time of the peptides, likely because of the low cell density required for this assay (data not shown). Therefore, the cell number of this assay was doubled to 80,000 cells/mL which gave a positive signal (Fig. 3B, D, and F). To validate that the peptides were still active at this 40-fold higher cell concentration than the initial assay (Table 1), we performed a viability assay. It is noteworthy that RR, D-RR, and WR at the same LD100 and time of death determined for the assay with 2,000 cells/mL retained their toxicity to forty times more cells than the original antimicrobial assay, causing cell viability loss of almost all cell population (Fig. S4). Our data clearly indicate that the bioinspired peptides induced hyperpolarization of their mitochondrial membrane potential leading to mitochondrial dysfunction in the tested yeasts. These results may exclude death as an apoptotic-like process because the loss of mitochondrial membrane potential is a hallmark of apoptosis [87]. Future studies should investigate the site of ROS production in the yeast mitochondria to gain a better understanding of how the stress triggered by the bioinspired peptides propagates within yeast cells, reaching and interfering with mitochondria.
Because ROS appear to be essential executors in regulated cell death with apoptotic-like phenotype in yeasts [64, 88] and our results pointed out ROS is linked to this process, we explored some regulated cell death phenotypes in yeasts. Indeed, apoptosis in fungi has been questioned recently [89]. However, it is undeniable that under certain conditions fungal cells die, and whose death exhibits phenotypic characteristics of intrinsic apoptosis in mammalian cells [65]. For this reason, we investigated some known markers of mammalian cell apoptosis to reinforce our results which indicate that a type of regulated cell death with phenotypic characteristics of mammalian cell apoptosis is being induced in C. tropicalis and C. albicans treated with the designed peptides, as suggested by Carmona-Gutierrez et al. [67]. We initially showed that the treated yeasts are under oxidative stress (Tables 2 and 3, Fig. S2), and exhibited a significant reduction in the cell size when treated with the designed peptides (Fig. 2). Both features are hallmarks of apoptosis in mammalian and yeast cells [48, 66]. We further evaluated whether the yeast metacaspases were activated upon peptides treatment using CaspACE FITC-VAD-FMK, a derivative of the pan-caspase inhibitor carbobenzoxyvalyl-alanyl-aspartyl-[O-methyl]-fluoromethylketone (Z-VAD-FMK), where the N-terminal blocking group was replaced by the fluorescein isothiocyanate (FITC) group. This replacement endowed the pan-caspase inhibitor with fluorescence and the ability to enter cells and bind activated caspases. Only C. tropicalis cells incubated with D-RR presented FITC-VAD-FMK positive cells as well as the positive control (AcA) (Figs. 4A, 5A, 6A) which was used to adjust such parameters as exposure time and excitation intensity. The activation of metacaspases in Candida by AcA, as well as other AMPs, is in accordance with other works [27, 90, 91] and also with the description of CaMAC1 gene which encodes a metacaspase in C. albicans [92], and therefore, support our results. The inhibition of metacaspase activation by AA in C. tropicalis incubated with D-RR (Fig. 6) provided a causal link between the protection of C. tropicalis cells from ROS production by this antioxidant (Table 2 and Fig. 2B). To gain a better comprehension of the role that metacaspase activation plays in the death induced by the bioinspired peptides, we treated yeast cells with the pan-caspase inhibitor Z-VAD-FMK. This inhibitor enters the cell and irreversibly binds to the catalytic site of active caspases, inhibiting their activity. Therefore, if this is the route of death, treatment with Z-VAD-FMK should protect the yeasts from the action of the peptides. We found the addition of 50 μM of the pan-caspase inhibitor in the antifungal assay did not protect Candida cells from the action of the bioinspired peptides (Figs. 4C, 5C, 6C). These results point out that the death induced by the designed peptides in C. tropicalis and C. albicans is metacaspase independent. Guaragnela et al. [93] studied S. cerevisiae death by AcA treatment and showed the activation of metacaspases by FITC-VAD-FMK and a non-protection from death by the pan-caspase inhibitor Z-VAD-FMK. Their results indicated that the death induced by AcA in the budding yeast was metacaspase independent. Later studies demonstrated that many factors can induce S. cerevisiae death independent of metacaspase activity [94]. The activation of metacaspases was demonstrated by FITC-VAD-FM for C. albicans treated with Rs-AFP2, and coincubation with Z-VAD-FMK halted Rs-AFP2-induced yeast death. However, the deletion of the CaMCA1 did not interfere with the peptide-induced death, indicating that other caspase-like proteases may be involved [90]. Therefore, our results may indicate that the death pathways activated by the designed peptides in yeast are metacaspase independent or there is another signal that once triggered commits cells to death as discussed above. Another regulated death with apoptosis-like features in mammalian cells analyzed in the present study was the detection of chromatin condensation by a fluorescent probe DAPI, which binds to the adenine and thymine-rich sequences of minor grooves of the DNA. Our results showed that the bioinspired peptides induce an alteration in the DAPI staining pattern (Figs. 4D, 5D, 6D). AMPs lead to both caspase-dependent [50] and independent [95] cell death with chromatin condensation. Due to the rapidity with which WR-induced death in C. albicans (Table 1), and RR-induced a metacaspase-independent cell death in C. tropicalis (Fig. 4A and C), we investigated whether the cell death was an accidental cell death (ACD) type caused by the bioinspired peptides by staining yeast cells with propidium iodide (PI). PI is a fluorescent probe that intercalates DNA and only enters cells with a ruptured cell membrane, being not permeable to the entire plasma membrane, and is excluded from live cells or cells undergoing early apoptosis, but binds to the nucleic acid of cells undergoing ACD and late apoptosis. RR and WR treatment resulted in positive PI labeling in C. tropicalis and C. albicans, respectively (Table 4), and negative labeling using D-RR and C. tropicalis (Table 4). AA had a protective effect on PI labeling in C. tropicalis incubated with RR; this result is in accordance with the treatment of C. tropicalis concomitantly with RR and AA in which C. tropicalis death was prevented (Table 2), and once again linked the toxic effect of RR on C. tropicalis to an uncontrolled ROS production. Additionally, the time of 15 min (Table 5) being the earliest that the PI labeling took place ruled out the possibility of death being late apoptosis. The same PI labeling time for C. tropicalis and C. albicans (Table 5) does not seem to be a mere coincidence and suggests the activation of a conserved mechanism by the peptides. Additionally, this result indicated that the fate of the yeasts after incubation with the peptides is being determined by the cells within this time interval of 15 min, and this finding will be further investigated in the future. In summary, for both RR and D-RR peptides, the induction of death in C. tropicalis was reversed by AA (Table 2), therefore suggesting that the death process is a regulated one. For RR and WR our results suggest an ACD once the cells treated with these peptides showed PI-positive labeling (Table 4). Although ACD is characterized by a well-defined feature, the augment in cell volume, in both mammalian and yeast cells [48, 66], our results indicated that Candida cells treated with RR and WR were PI positive and were undergoing cell shrinkage (Fig. 2). The permeabilization of the yeast membranes allowed the efflux of ions, including K+, and if K+ leakage was the case, it might explain the shrinkage and not cell volume expansion of the treated yeasts [96, 97].
Conclusions
In this study, we have gleaned information from our previous work on the amino acid residues in the γ-core region of plant defensins important for their biological activity [29]. Next, we designed the RR, D-RR, and WR peptides which exhibited improved antimicrobial activity against opportunistic yeasts and low toxicity to mouse macrophages and human monocytes compared to the original peptide [29]. Herein, we report the effects of these bioinspired peptides on Candida yeasts and propose a mechanism of action in regard to distinct types of induced cell death (Fig. 7). We showed that exposure of C. tropicalis to RR and D-RR promoted a regulated accidental death (Fig. 7) and metacaspase-independent regulated cell death, respectively (Fig. 7), while WR promoted ACD in C. albicans (Fig. 7). Our results were obtained with the LD100 and within the time that the peptides induce the yeast death. Within this temporal frame, our results allow us to gain clarity on the events triggered by the peptide-cell interaction and their temporal order, providing a better understanding of the death process induced by these peptides. Future studies should provide further details of the action time course. The understanding of the process of fungal cell death triggered by AMP interaction is important to support the clinical use of antifungal molecules, since the lack of understanding of the mechanism of action leads to failure in more advanced clinical trials. Our results can be mirrored by other AMPs and derived peptides helping to pave the way for the clinical use of these molecules. Moreover, the understanding of the mode of action of bioinspired peptides is important in light of the fact that regulated cell death pathways are also involved in Candida virulence [98] raising the possibility to manipulate these pathways for antifungal therapy purposes.
Schematic depiction of the proposed model describing the mechanism of action of RR and D-RR on Candida tropicalis, and WR on Candida albicans. Based on the results present study using probes to cellular process (ROS, mitochondrial hyperpolarization, cell size reduction/vacuolization, chromatin condensation, accidental cell death, metacaspase activation) and inhibitors (ascorbic acid, metacaspase inhibitor) allowed to differentiate the type of cell death promoted by each peptide
Data availability
All relevant data are within the manuscript and supplementary information files. Data will be shared upon request with the corresponding author.
Abbreviations
- AA:
-
L-ascorbic acid
- AcA:
-
Acetic acid
- ACD:
-
Accidental cell death
- AMP:
-
Antimicrobial peptides
- CoQ:
-
Coenzyme Q
- CFU:
-
Colony forming units
- DAPI:
-
4’,6-Diamidino-2-phenylindole
- DIC:
-
Differential interference contrast
- DMSO:
-
Dimethyl sulfoxide
- D-RR:
-
D-A36,42,44R37,38γ32-46VuDef
- ETC:
-
Electron transport chain
- GSH:
-
γ-L-glutamyl-L-cysteinyl-glycine
- OH• :
-
Hydroxyl anion
- H2DCFDA:
-
2’,7’-Dichlorodihydrofluorescein diacetate
- ∆ψm :
-
Mitochondrial membrane potential
- O2 :
-
Molecular oxygen
- NAC:
-
N-acetyl-L-cysteine
- nd:
-
Not determined
- PI:
-
Propidium iodide
- RET:
-
Reverse electron transport
- ROS:
-
Reactive oxygen species
- RR:
-
A36,42,44R37,38γ32-46VuDef
- WR:
-
A42,44R37,38W36,39γ32-46VuDef
- SD:
-
Standard deviation
References
Medina E, Pieper DH (2016) Tackling threats and future problems of multidrug-resistant bacteria. Curr Top Microbiol Immunol 398:3–33. https://doi.org/10.1007/82_2016_492/COVER
Bartoletti M, Giannella M, Tedeschi SE, Viale P (2018) Multidrug-resistant bacterial infections in solid organ transplant candidates and recipients. Infect Dis Clin North Am 32:551–580. https://doi.org/10.1016/J.IDC.2018.04.004
Browne K, Chakraborty S, Chen R, Willcox MD, Black DS, Walsh WR, Kumar N (2020) A new era of antibiotics: the clinical potential of antimicrobial peptides. Int J Mol Sci 21:1–23. https://doi.org/10.3390/ijms21197047
Ferri M, Ranucci E, Romagnoli PE, Giaccone V (2017) Antimicrobial resistance: a global emerging threat to public health systems. Crit Rev Food Sci Nutr 57:2857–2876. https://doi.org/10.1080/10408398.2015.1077192
Pham TN, Loupias P, Dassonville-Klimpt A, Sonnet P (2019) Drug delivery systems designed to overcome antimicrobial resistance. Med Res Rev 39:2343–2396. https://doi.org/10.1002/MED.21588
Lorch JM, Meteyer CU, Behr MJ, Boyles JG, Cryan PM, Hicks AC, Ballmann AE, Coleman JTH et al (2011) Experimental infection of bats with Geomyces destructans causes white-nose syndrome. Nature 480:376–378. https://doi.org/10.1038/nature10590
Fisher MC, Garner TWJ (2020) Chytrid fungi and global amphibian declines. Nat Rev Microbiol 18:332–343. https://doi.org/10.1038/s41579-020-0335-x
Boyles JG, Cryan PM, McCracken GF, Kunz TH (2011) Conservation: economic importance of bats in agriculture. Science 332:41–42. https://doi.org/10.1126/SCIENCE.1201366
Colón-Gaud C, Whiles MR, Kilham SS, Lips KR, Pringle CM, Connelly S, Peterson SD (2009) Assessing ecological responses to catastrophic amphibian declines: patterns of macroinvertebrate production and food web structure in upland Panamanian streams. Limnol Oceanogr 54:331–343. https://doi.org/10.4319/LO.2009.54.1.0331
Fisher MC, Hawkins NJ, Sanglard D, Gurr SJ (2018) Worldwide emergence of resistance to antifungal drugs challenges human health and food security. Science 360:739–742. https://doi.org/10.1126/SCIENCE.AAP7999
Food and Agriculture Organization of the United Nations (2017) Strategic work of FAO for sustainable food and agriculture. 6488EN/1/01.17
Ashraf N, Kubat RC, Poplin V, Adenis AA, Denning DW, Wright L, McCotter O, Schwartz IS, Jackson BR, Chiller T, Bahr NC (2020) Re-drawing the maps for endemic mycoses. Mycopathologia 185:843–865. https://doi.org/10.1007/S11046-020-00431-2
Rodrigues ML, Nosanchuk JD (2020) Fungal diseases as neglected pathogens: a wake-up call to public health officials. PLoS Negl Trop Dis 14:e0007964. https://doi.org/10.1371/JOURNAL.PNTD.0007964
Pristov KE, Ghannoum MA (2019) Resistance of Candida to azoles and echinocandins worldwide. Clin Microbiol Infect 25:792–798. https://doi.org/10.1016/J.CMI.2019.03.028
Garcia-Rubio R, Mellado C-E, E, (2017) Triazole resistance in Aspergillus species: an emerging problem. Drugs 77:599–613. https://doi.org/10.1007/S40265-017-0714-4
Forsberg K, Woodworth K, Walters M, Berkow EL, Jackson B, Chiller T, Vallabhaneni S (2019) Candida auris: the recent emergence of a multidrug-resistant fungal pathogen. Med Mycol 57:1–12. https://doi.org/10.1093/MMY/MYY054
Lockhart SR, Etienne KA, Vallabhaneni S, Farooqi J, Chowdhary A, Govender NP et al (2017). Simultaneous emergence of multidrug-resistant Candida auris on 3 continents confirmed by whole-genome sequencing and epidemiological analyses. Clin Infect Dis 64:134–140.https://doi.org/10.1093/CID/CIW691
Goel RR, Apostolidis SA, Painter MM, Mathew D, Pattekar A, Kuthuru O, Gouma S, Hicks P et al (2021) Distinct antibody and memory B cell responses in SARSCoV-2 naïve and recovered individuals following mRNA vaccination. Sci Immunol 6:1–19. https://doi.org/10.1126/sciimmunol.abi6950
Hoque MN, Akter S, Mishu ID, Islam MR, Rahman MS, Akhter M, Islam I, Hasan MM et al (2021) Microbial co-infections in COVID-19: associated microbiota and underlying mechanisms of pathogenesis. Microb Pathog 156:104941. https://doi.org/10.1016/J.MICPATH.2021.104941
Martins AC, Psaltikidis EM, Lima TC, Fagnani R, Schreiber AZ, Conterno LO, Kamei K et al (2021) COVID-19 and invasive fungal coinfections: a case series at a Brazilian referral hospital. J Mycol Med 31:101175. https://doi.org/10.1016/J.MYCMED.2021.101175
Rezasoltani S, Yadegar A, Hatami B, Aghdaei HA, Zali MR (2020) Antimicrobial resistance as a hidden menace lurking behind the COVID-19 Outbreak: the global impacts of too much hygiene on AMR. Front Microbiol 11:3097. https://doi.org/10.3389/FMICB.2020.590683
Ansari S, Hays JP, Kemp A, Okechukwu R, Murugaiyan J, Ekwanzala MD, Alvarez MJR, Paul-Satyaseela M et al (2021) The potential impact of the COVID-19 pandemic on global antimicrobial and biocide resistance: an AMR Insights global perspective. JAC Antimicrob Resist 3:dblab038. https://doi.org/10.1093/JACAMR/DLAB038
Knight GM, Glover RE, McQuaid CF, Olaru ID, Gallandat K, Leclerc QJ, Fuller NM et al (2021) Antimicrobial resistance and COVID-19: Intersections and implications. Elife 10:1–27. https://doi.org/10.7554/ELIFE.64139
Rusic D, Vilovic M, Bukic J, Leskur D, Perisin AS, Kumric M, Martinovic D, Petric A, Modun D, Bozic J (2021) Implications of COVID-19 pandemic on the emergence of antimicrobial resistance: adjusting the response to future outbreaks. Elife 11:220. https://doi.org/10.3390/LIFE11030220
Moretta A, Scieuzo C, Petrone AM, Salvia R, Manniello MD, Franco A, Lucchetti D, Vassallo A, Vogel H, Sgambato A, Falabella P (2021) Antimicrobial peptides: a new hope in biomedical and pharmaceutical fields. Front Cell Infect Microbiol 11:668632. https://doi.org/10.3389/FCIMB.2021.668632
Li X, Zuo S, Wang B, Zhang K, Wang Y (2022) Antimicrobial mechanisms and clinical application prospects of antimicrobial peptides. Molecules 27:2675. https://doi.org/10.3390/MOLECULES27092675
Mello ÉO, Taveira GB, Carvalho AO, Gomes VM (2019) Improved smallest peptides based on positive charge increase of the γ-core motif from PνD1 and their mechanism of action against Candida species. Int J Nanomedicine 14:407–420. https://doi.org/10.2147/IJN.S187957
Souza GS, Carvalho LP, Melo EJT, Silva FCV, Machado OLT, Gomes VM, Carvalho AO (2019) A synthetic peptide derived of the β2–β3 loop of the plant defensin from Vigna unguiculata seeds induces Leishmania amazonensis apoptosis-like cell death. Amino Acids 51:1633–1648. https://doi.org/10.1007/S00726-019-02800-8
Toledo EB, Lucas DR, Simão TLBV, Calixto SD, Lassounskaia E, Muzitano MF, Damica FZ, Gomes VM, Carvalho AO (2021) Design of improved synthetic antifungal peptides with targeted variations in charge, hydrophobicity and chirality based on a correlation study between biological activity and primary structure of plant defensin γ-cores. Amino Acids 53:219–237. https://doi.org/10.1007/S00726-020-02929-X
Schäfer AB, Wenzel M (2020) A how-to guide for mode of action analysis of antimicrobial peptides. Front Cell Infect Microbiol 10:540898. https://doi.org/10.3389/FCIMB.2020.540898
Soares JR, Melo JTM, da Cunha M, Fernandes KVS, Taveira GB, Silva Pereira L et al (2017) Interaction between the plant ApDef1 defensin and Saccharomyces cerevisiae results in yeast death through a cell cycle- and caspase-dependent process occurring via uncontrolled oxidative stress. Biochim Biophys Acta Gen Subj 1861:3429–3443. https://doi.org/10.1016/J.BBAGEN.2016.09.005
Grosfeld EV, Bidiuk VA, Mitkevich OV, Ghazy ESMO, Kushnirov VV, Alexandrov AI (2021) A systematic survey of characteristic features of yeast cell death triggered by external factors. J Fungi 7:886. https://doi.org/10.3390/JOF7110886
Berridge MV, Tan HPM, AS, (2005) Tetrazolium dyes as tools in cell biology: new insights into their cellular reduction. Biotechnol Annu Rev 11:127–152. https://doi.org/10.1016/S1387-2656(05)11004-7
Tsukatani T, Oba T, Ukeda H, Matsumoto K (2003) Spectrophotometric assay of yeast vitality using 2,3,5,6-tetramethyl-1,4-benzoquinone and tetrazolium salts. Anal Sci 19:659–664. https://doi.org/10.2116/ANALSCI.19.659
Barreto-Santamaría A, Patarroyo ME, Curtidor H (2019) Designing and optimizing new antimicrobial peptides: all targets are not the same. Crit Rev Clin Lab Sci 56:351–373. https://doi.org/10.1080/10408363.2019.1631249
Torres MDT, Sothiselvam S, Lu TK, de la Fuente-Nunez C (2019) Peptide design principles for antimicrobial applications. J Mol Biol 431:3547–3567. https://doi.org/10.1016/j.jmb.2018.12.015
Thevissen K, Terras FRG, Broekaert WF (1999) Permeabilization of fungal membranes by plant defensins inhibits fungal growth. Appl Environ Microbiol 65:5451–5458. https://doi.org/10.1128/AEM.65.12.5451-5458.1999
Schmitt P, Rosa RD, Destoumieux-Garzón D (2016) An intimate link between antimicrobial peptide sequence diversity and binding to essential components of bacterial membranes. Biochim Biophys Acta (BBA) - Biomembr 1858:958–970. https://doi.org/10.1016/J.BBAMEM.2015.10.011
Chang CK, Lan KMC, CY, (2021) Antimicrobial activity of the peptide LfcinB15 against Candida albicans. J fungi 7:519. https://doi.org/10.3390/JOF7070519
Park CB, Kim HS, Kim SC (1998) Mechanism of action of the antimicrobial peptide buforin II: buforin II kills microorganisms by penetrating the cell membrane and inhibiting cellular functions. Biochem Biophys Res Commun 244:253–257. https://doi.org/10.1006/BBRC.1998.8159
Aerts AM, François IEJA, Meert EMK, Li QT, Cammue BPA, Thevissen K (2007) The antifungal activity of RsAFP2, a plant defensin from Raphanus sativus, involves the induction of reactive oxygen species in Candida albicans. Microb Physiol 13:243–247. https://doi.org/10.1159/000104753
Mello EO, Ribeiro SFF, Carvalho AO, Santos IS, da Cunha M, Santa-Catarina C, Gomes VM (2011) Antifungal activity of PvD1 defensin involves plasma membrane permeabilization, inhibition of medium acidification, and induction of ROS in fungi cells. Curr Microbiol 62:1209–1217. https://doi.org/10.1007/S00284-010-9847-3
He J, Krauson AJ, Wimley WC (2014) Toward the de novo design of antimicrobial peptides: lack of correlation between peptide permeabilization of lipid vesicles and antimicrobial, cytolytic, or cytotoxic activity in living cells. Peptide Sci 102:1–6. https://doi.org/10.1002/BIP.22281
Muñoz A, Gandía M, Harries E, Carmona L, Read ND, Marcos JF (2013) Understanding the mechanism of action of cell-penetrating antifungal peptides using the rationally designed hexapeptide PAF26 as a model. Fungal Biol Rev 26:146–155. https://doi.org/10.1016/J.FBR.2012.10.003
Peschel A, Jack RW, Otto M, Collins LV, Staubitz P, Nicholson G, Kalbacher H, Nieuwenhuizen WF, Jung G et al (2001) Staphylococcus aureus resistance to human defensins and evasion of neutrophil killing via the novel virulence factor MprF is based on modification of membrane lipids with L-lysine. Exp Med 193:1067–1076. https://doi.org/10.1084/JEM.193.9.1067
Abdi M, Mirkalantari S, Amirmozafari N (2019) Bacterial resistance to antimicrobial peptides. J Pept Sci 25e3210. https://doi.org/10.1002/PSC.3210
Kusch H, Engelmann S, Albrecht D, Morschhäuser J, Hecker M (2007) Proteomic analysis of the oxidative stress response in Candida albicans. Proteomics 7:686–697. https://doi.org/10.1002/PMIC.200600575
Bortner CD, Cidlowski JA (2020) Ions, the movement of water and the apoptotic volume decrease. Front Cell Dev Biol 8:611211. https://doi.org/10.3389/FCELL.2020.611211
Arrigoni O, de Tullio MC (2002) Ascorbic acid: much more than just an antioxidant. Biochim Biophys Acta Gen Subj 1569:1–9. https://doi.org/10.1016/S0304-4165(01)00235-5
Lee W, Lee DG (2017) Reactive oxygen species modulate itraconazole-induced apoptosis via mitochondrial disruption in Candida albicans. Free Radic Res 52:39–50. https://doi.org/10.1080/10715762.2017.1407412
Hayes BME, Bleackley MR, Wiltshire JL, Anderson MA, Traven A, van der Weerden NL (2013) Identification and mechanism of action of the plant defensin NaD1 as a new member of the antifungal drug arsenal against Candida albicans. Antimicrob Agents Chemother 57:3667–3675. https://doi.org/10.1128/AAC.00365-13
Guaragnella N, Passarella S, Marra E, Giannattasio S (2010) Knock-out of metacaspase and/or cytochrome c results in the activation of a ROS-independent acetic acid-induced programmed cell death pathway in yeast. FEBS Lett 584:3655–3660. https://doi.org/10.1016/J.FEBSLET.2010.07.044
Bullock CB, McNabb DS, Pinto I (2020) Whole-genome approach to understanding the mechanism of action of a histatin 5-derived peptide. Antimicrob Agents Chemother 64:e01698-e1719. https://doi.org/10.1128/AAC.01698-19
Machida K, Tanaka T (1999) Farnesol-induced generation of reactive oxygen species dependent on mitochondrial transmembrane potential hyperpolarization mediated by F0F1-ATPase in yeast. FEBS Lett 462:108–112. https://doi.org/10.1016/S0014-5793(99)01506-9
Andrés MT, Viejo-Díaz M, Fierro JF (2008) Human lactoferrin induces apoptosis-like cell death in Candida albicans: critical role of K+-channel-mediated K+ efflux. Antimicrob Agents Chemother 52:4081–4088. https://doi.org/10.1128/AAC.01597-07
Ojha R, Khan L, Manzoor N (2009) Ascorbic acid modulates pathogenecity markers of Candida albicans. Int J Microbiol Res 1:19–24. https://doi.org/10.9735/0975-5276.1.1.19-24
Costa V, Moradas-Ferreira P (2001) Oxidative stress and signal transduction in Saccharomyces cerevisiae: insights into ageing, apoptosis and diseases. Mol Aspects Med 22:217–246. https://doi.org/10.1016/S0098-2997(01)00012-7
Rinnerthaler M, Büttner S, Laun P, Heeren G, Felder TK, Klinger H, Weinberger M et al (2012) Yno1p/Aim14p, a NADPH-oxidase ortholog, controls extramitochondrial reactive oxygen species generation, apoptosis, and actin cable formation in yeast. Proc Natl Acad Sci USA 109:8658–8663. https://doi.org/10.1073/PNAS.1201629109
Schippers JHM, Nguyen HM, Lu D, Schmidt R, Mueller-Roeber B (2012) ROS homeostasis during development: an evolutionary conserved strategy. Cell Mol Life Sci 69:3245–3257. https://doi.org/10.1007/S00018-012-1092-4
Aruoma OI, Halliwell B, Hoey BM, Butler J (1989) The antioxidant action of N-acetylcysteine: its reaction with hydrogen peroxide, hydroxyl radical, superoxide, and hypochlorous acid. Free Radic Biol Med 6:593–597. https://doi.org/10.1016/0891-5849(89)90066-X
Aldini G, Altomare A, Baron G, Vistoli G, Carini M, Borsani L, Sergio F (2018) N-Acetylcysteine as an antioxidant and disulphide breaking agent: the reasons why. Free Radic Res 52:751–762. https://doi.org/10.1080/10715762.2018.1468564
Pócsi I, Prade RA, Penninckx MJ (2004) Glutathione, altruistic metabolite in fungi. Adv Microb Physiol 49:1–76. https://doi.org/10.1016/S0065-2911(04)49001-8
Toledano MB, Delaunay-Moisan A, Outten CE, Igbaria A (2013) Functions and cellular compartmentation of the thioredoxin and glutathione pathways in yeast. Antioxid Redox Signal 18:699–1711. https://doi.org/10.1089/ARS.2012.5033
Kumar C, Igbaria A, D’Autreaux B, Planson AG, Junot C, Godat E, Bachhawat AK, Delaunay-Moisan A, Toledano MB (2011) Glutathione revisited: a vital function in iron metabolism and ancillary role in thiol-redox control. EMBO J 30:2044–2056. https://doi.org/10.1038/EMBOJ.2011.105
Madeo F, Fröhlich E, Ligr M, Grey M, Sigrist SJ, Wolf DH, Fröhlich KU (1999) Oxygen stress: a regulator of apoptosis in yeast. J Cell Biol 145:757–767. https://doi.org/10.1083/JCB.145.4.757
Chaves SR, Rego A, Martins VM, Santos-Pereira C, Sousa MJ, Côrte-Real M (2021) Regulation of cell death induced by acetic acid in yeasts. Front Cell Dev Biol 9:642375. https://doi.org/10.3389/FCELL.2021.642375
Carmona-Gutierrez D, Bauer MA, Zimmermann A, Aguilera A, Austriaco N, Ayscough K, Balzan R, Bar-Nun S, Barrientos A et al (2018) Guidelines and recommendations on yeast cell death nomenclature. Microbial Cell 5:4–31. https://doi.org/10.15698/MIC2018.01.607
Walker GM (1999) Synchronization of yeast cell populations. Methods Cell Sci 21:87–93. https://doi.org/10.1023/A:1009824520278
Yun JE, Lee DG (2017) Role of potassium channels in chlorogenic acid-induced apoptotic volume decrease and cell cycle arrest in Candida albicans. Biochim Biophys Acta Gen Subj 1861:585–592. https://doi.org/10.1016/J.BBAGEN.2016.12.026
Baev D, Li XS, Dong J, Keng P, Edgerton M (2002) Human salivary histatin 5 causes disordered volume regulation and cell cycle arrest in Candida albicans. Infect Immun 70:4777–4784. https://doi.org/10.1128/IAI.70.9.4777-4784.2002
Kwon YY, Choi KM, Cho CY, Lee CK (2015) Mitochondrial efficiency-dependent viability of Saccharomyces cerevisiae mutants carrying individual electron transport chain component deletions. Mol Cells 38:1054–1063. https://doi.org/10.14348/MOLCELLS.2015.0153
Zhao RZ, Jiang S, Zhang L, Yu Z (2019) Mitochondrial electron transport chain, ROS generation and uncoupling. Int J Mol Med 44(3–15):72. https://doi.org/10.3892/IJMM.2019.4188/HTML
Venditti P, di Stefano L, di Meo S (2013) Mitochondrial metabolism of reactive oxygen species. Int J Mol Med Adv Sci 13:71–82. https://doi.org/10.1016/J.MITO.2013.01.008
Mailloux RJ (2015) Teaching the fundamentals of electron transfer reactions in mitochondria and the production and detection of reactive oxygen species. Redox Biol 4:381–398. https://doi.org/10.1016/J.REDOX.2015.02.001
Napolitano G, Fasciolo G, Venditti P (2021) Mitochondrial management of reactive oxygen species. antioxidants 10:1824. https://doi.org/10.3390/ANTIOX10111824
Robb EL, Hall AR, Prime TA, Eaton S, Szibor M, Viscomi C, James AM, Murphy MP (2018) Control of mitochondrial superoxide production by reverse electron transport at complex I. J Biol Chem 293:9869–9879. https://doi.org/10.1074/JBC.RA118.003647
Milani G, Jarmuszkiewicz W, Sluse-Goffart CM, Schreiber AZ, Vercesi AE, Sluse FE (2001) Respiratory chain network in mitochondria of Candida parapsilosis: ADP/O appraisal of the multiple electron pathways. FEBS Lett 508:231–235. https://doi.org/10.1016/S0014-5793(01)03060-5
Ruy F, Vercesi AE, Kowaltowski AJ, Ruy F, Vercesi AE, Kowaltowski AJ (2006) Inhibition of specific electron transport pathways leads to oxidative stress and decreased Candida albicans proliferation. J Bioenerg Biomem 38(29–135):78. https://doi.org/10.1007/S10863-006-9012-7
Castro A, Lemos C, Falcão A, Glass NL, Videira A (2008) Increased resistance of complex I mutants to phytosphingosine-induced programmed cell death. J Biol Chem 283:19314–19321. https://doi.org/10.1074/JBC.M802112200
Li D, Chen H, Florentino A, Alex D, Sikorski P, Fonzi WA, Calderone R (2011) Enzymatic dysfunction of mitochondrial complex I of the Candida albicans goa1 mutant is associated with increased reactive oxidants and cell death. Eukaryot Cell 10:672–68280. https://doi.org/10.1128/EC.00303-10
Xu Y, Wang Y, Yan L, Liang RM, di Dai B, Tang RJ, Gao PH, Jiang YY (2009) Proteomic analysis reveals a synergistic mechanism of fluconazole and berberine against fluconazole-resistant Candida albicans: endogenous ROS augmentation. J Proteome Res 8:5296–5304. https://doi.org/10.1021/PR9005074
Wu XZ, Cheng AX, Sun LM, Sun SJ, Lou HX (2009) Plagiochin E, an antifungal bis(bibenzyl), exerts its antifungal activity through mitochondrial dysfunction-induced reactive oxygen species accumulation in Candida albicans. Biochim Biophys Acta (BBA) - Gen Subj 1790:770–777. https://doi.org/10.1016/J.BBAGEN.2009.05.002
Ramzan R, Michels S, Weber P, Rhiel A, Irqsusi M, Rastan AJ, Culmsee C, Vogt S (2019) Protamine sulfate induces mitochondrial hyperpolarization and a subsequent increase in reactive oxygen species production. J Pharmacol Exp Ther 370:308–317. https://doi.org/10.1124/JPET.119.257725
Skulachev VP (2006) Bioenergetic aspects of apoptosis, necrosis and mitoptosis. Apoptosis 11:473–485. https://doi.org/10.1007/S10495-006-5881-9
Pozniakovsky AI, Knorre DA, Ov M, Hyman AA, Skulachev VP, Severin FF (2005) Role of mitochondria in the pheromone- and amiodarone-induced programmed death of yeast. J Cell Biol 168:257–269. https://doi.org/10.1083/JCB.200408145
Gabaldón T, Rainey D, Huynen MA (2005) Tracing the evolution of a large protein complex in the eukaryotes, NADH:ubiquinone oxidoreductase (Complex I). J Mol Biol 348:857–870. https://doi.org/10.1016/J.JMB.2005.02.067
Guaragnella N, Ždralevic M, Antonacci L, Passarella S, Marra E, Giannattasio S (2012) The role of mitochondria in yeast programmed cell death. Front Oncol 2:70. https://doi.org/10.3389/FONC.2012.00070
Madeo F, Herker E, Maldener C, Wissing S, Lächelt S, Herlan M, Fehr M, Lauber K, Sigrist SJ, Wesselborg S, Fröhlich KU (2002) A caspase-related protease regulates apoptosis in yeast. Mol Cell 9:911–917. https://doi.org/10.1016/S1097-2765(02)00501-4
Hardwick JM (2018) Do fungi undergo apoptosis-like programmed cell death? mBio 9:e00948–18. https://doi.org/10.1128/MBIO.00948-18
Aerts AM, Carmona-Gutierrez D, Lefevre S, Govaert G, François IEJA, Madeo F, Santos R, Cammue BPA, Thevissen K (2009) The antifungal plant defensin RsAFP2 from radish induces apoptosis in a metacaspase independent way in Candida albicans. FEBS Lett 583:2513–2516. https://doi.org/10.1016/J.FEBSLET.2009.07.004
Taveira GB, Mello ÉO, Souza SB, Monteiro RM, Ramos AC, Carvalho AO, Rodrigues R, Okorokov LA, Gomes VM (2018) Programmed cell death in yeast by thionin-like peptide from Capsicum annuum fruits involving activation of caspases and extracellular H+ flux. Biosci Rep 38:BSR20180119. https://doi.org/10.1042/BSR20180119/57557
Cao YY, Huang S, di Dai B, Zhu ZY, Lu H, Dong LL, Cao YB, Wang Y, Gao PH, Chai YF, Jiang YY (2009) Candida albicans cells lacking CaMCA1-encoded metacaspase show resistance to oxidative stress-induced death and change in energy metabolism. Fungal Genet Biol 46:183–189. https://doi.org/10.1016/J.FGB.2008.11.001
Guaragnella N, Pereira C, Sousa MJ, Antonacci L, Passarella S, Côrte-Real M, Marra E, Giannattasio S (2006) YCA1 participates in the acetic acid induced yeast programmed cell death also in a manner unrelated to its caspase-like activity. FEBS Lett 580:6880–6884. https://doi.org/10.1016/J.FEBSLET.2006.11.050
Liang Q, Li W, Zhou B (2008) Caspase-independent apoptosis in yeast. Biochim Biophys Acta 1783:1311–1319. https://doi.org/10.1016/J.BBAMCR.2008.02.018
Morton CO, dos Santos SC, Coote P (2007) An amphibian-derived, cationic, α-helical antimicrobial peptide kills yeast by caspase-independent but AIF-dependent programmed cell death. Mol Microbiol 65:494–507. https://doi.org/10.1111/J.1365-2958.2007.05801.X
Xu Y, Ambudkar I, Yamagishi H, Swaim W, Walsh TJ, O’Connell BC (1999) Histatin 3-mediated killing of Candida albicans: effect of extracellular salt concentration on binding and internalization. Antimicrob Agents Chemother 43:2256–2262. https://doi.org/10.1128/AAC.43.9.2256
Lee H, Woo ER, Lee DG (2018) Apigenin induces cell shrinkage in Candida albicans by membrane perturbation. FEMS Yeast Res 18:foy003. https://doi.org/10.1093/FEMSYR/FOY003
Nam M, Kim SH, Jeong JH, Kim S, Kim J (2022) Roles of the pro-apoptotic factors CaNma111 and CaYbh3 in apoptosis and virulence of Candida albicans. Sci Rep 12:1–9. https://doi.org/10.1038/s41598-022-11682-y
Acknowledgements
The work was supported by the Brazilian agencies Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq), Fundação de Amparo à Pesquisa do Estado do Rio de Janeiro (FAPERJ, process nº. E-26/202.760/2018-Bolsa), and Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (Finance Code 001) as well as the Universidade Estadual do Norte Fluminense Darcy Ribeiro. We acknowledge Luiz Carlos Dutra de Souza and Valeria Miguelote Kokis for their technical support.
Author information
Authors and Affiliations
Contributions
D.R.L., F.Z.D., and E.B.T.: investigation, methodology, validation, writing—review and editing. A.J.D.C. and A.L.O.F.: investigation, writing—review and editing. V.M.G. investigation, A.O.C.: conceptualization, methodology, supervision, project administration, funding acquisition, writing—original draft.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Highlights
• There is a need to develop efficacious therapeutics for the treatment of fungal diseases.
• ROS induction by bioinspired peptides was linked to mitochondria hyperpolarization.
• ROS are not the final yeast death executor and a second signal seems to be operating.
• Cell fate of yeasts is determined after 15 min of interaction with the peptides.
• Different death mechanisms are induced by three bioinspired peptides against yeasts.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Lucas, D.R., Damica, F.Z., Toledo, E.B. et al. Bioinspired peptides induce different cell death mechanisms against opportunistic yeasts. Probiotics & Antimicro. Prot. 16, 649–672 (2024). https://doi.org/10.1007/s12602-023-10064-8
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12602-023-10064-8