Abstract
Biological invasions are one of the major constraints worldwide because of the economic and environmental consequences they may pose. Root mealybugs (Hemiptera: Coccomorpha: Rhizoecidae) are continuously transported worldwide with plant soil because of their cryptic behavior. In this study it was reported the presence of Ripersiella multiporifera Jansen in Sicily for the first time and discussed the presence of R. maasbachi (Jansen) and R. hibisci (Kawai & Takagi) in Italy. A key is provided to identify the species of Rhizoecidae currently known in Italy. The species were identified by morphological features and characterized molecularly by using a partial COI mitochondrial gene. Our report alerts the presence of alien root mealybugs in Italy. R. hibisci is a quarantine species listed in Annex IIA of EU Regulation 2019/2072.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Exotic insects are continually introduced every year in countries as a result of the trade and transport of plants and plant material with potential threats to biodiversity in natural areas and/or risk for economically relevant crops (Perrings et al., 2002; Riegler, 2018). This phenomenon increased in the last century and it has been exacerbated by the current climate change and global transporting as well (Zeng et al., 2020). Quarantine regulations and national programs are generally established to prevent the movement of pests among the regions and this objective is commonly achieved by the use of pest-and pathogen-free plant materials (Schrader & Unger, 2003).
In Italy, the number of introduced alien insects increases every year, 57.7% of them are Hemiptera of which 97% are Sternorrhyncha (Jucker & Lupi, 2011). Inghilesi et al. (2013) showed that in Italy and Europe, Sternorrhyncha represent the highest percentage of exotic insect orders compared with the number of species in the world. Most of these insects have been introduced through the trade and movement of ornamental plants. The scale insects (Hemiptera: Coccomorpha), due to their morphology, biology and behavior, are mainly concealed on hidden parts of host plants that contribute largely to their dispersion (Mazzeo et al., 2014, 2020), especially when they live on roots. Among scale insects, root mealybugs are common but difficult to detect and identify with the naked eye due to their small size and cryptic behavior.
The family Rhizoecidae (Hemiptera: Coccomorpha), recently instituted by Hodgson (2012), includes 19 genera and 238 species of “root mealybugs” or “ground mealybugs”, living and feeding on plant rootlets (Kozár & Konczné Benedicty, 2007; Kaydan et al., 2019). Within this family, the genus Ripersiella has changed its status several times, being sometimes considered as a synonym of the very close genus Rhizoecus. Recently Szita et al. (2020) discussed the position of the two genera and, following many other authors, considered Ripersiella as a distinct genus (Kozár & Konczné Benedicty, 2007). Ripersiella is characterized by the presence of bitubular pores on the dorsum and venter, antennae 5 or 6 segmented, legs well developed, and anal ring bearing 6 setae (Kozár & Konczné Benedicty, 2007).
All the species of Ripersiella live in the soil. However, it is hard to understand which plant they feed because specimens are often observed loosely in the soil and/or found on the roots of many plant species, as they can be collected only by sifting leaf litter and/or sieving soil samples (Kaydan et al., 2018, 2019).
The species of Ripersiella occur preferably in humid soils even if some of them prefer xerophilous habitats (Kozár & Konczné Benedicty, 2007). Some of them can perform more than one generation per year as the life cycle is influenced by the temperature and host plant. They are bisexual and females produce eggs in white, loose, and waxy ovisacs (EFSA PLH PANEL, 2020; Jansen and Westenberg, 2015).
In the last year, the presence of Ripersiella species has been detected by Phytosanitary Service in Sicily (Italy) during inspections of ornamental plants in commercial nurseries. Almost all the specimens collected belonged to Ripersiella hibisci (Kawai & Takagi, 1971) (Fig. 1), a species listed in Annex IIA of EU Regulation 2019/2072 as confirmed by European Food and Safety Agency (EFSA PLH Panel, 2020) and intercepted in Italy in the past (Mazzeo et al., 2014). Following the detection of R. hibisci, the Regional Phytosanitary Service has adopted the phytosanitary prescriptions aimed to eradicate outbreaks of this mealybug pursuant to Regional Decree n. 2794 of 2021/07/23: official controls in nurseries in Sicily were increased; the professional operator’s authorization to issue plant passports for R. hibisci host species is suspended until the outbreak has been eradicated; the infested plots have been isolated, immobilized and subjected to insecticide treatments or eliminated. The European Plant Protection Organism (EPPO, 2022b) considered R. hibisci eradicated in all sites in December 2021, but new records during 2022 and in early 2023 in Sicily (Table 1) led Regional Phytosanitary Service to apply specific measures for mealybug eradication.
During the monitoring in ornamental plant nurseries of Eastern Sicily, several mealybugs have been found on the roots of Sansevieria trifasciata Prain (Asparagaceae) and Camellia sp. (Theaceae). The specimens collected from S. trifasciata have been identified as Ripersiella multiporifera Jansen, 2008, a species that was for the first time intercepted and described by Jansen (2008) in the Netherlands. The specimens collected on Camellia sp. have been identified as Ripersiella maasbachi (Jansen, 2003) and promptly reported to EPPO (EPPO, 2022a). Ripersiella maasbachi was described by Jansen (2003) in the Netherlands, where the species was regularly intercepted on Sageretia sp. (Rhamnaceae) bonsai and other plants imported from China (Jansen, 2003). In this study, R. multiporifera, R. maasbachi and R. hibisci are reported and a taxonomic key of Rhizoecidae recorded in Italy is presented.
Materials and methods
The specimens were collected from soil and roots inside potted plants found in the ornamental plant nurseries in Eastern Sicily between 2021 and 2023 (Table 1). The living adult females were preserved in 70% alcohol solution in plastic vials. The specimens were then prepared for morphological identification and molecular characterization.
Morphological identification
Adult females were identified using a compound microscope; the adult females were slide-mounted in Canada balsam using the method described by Williams and Watson (1988). In particular, the females were cleaned and clarified by passages in 10% KOH, ethanol (70% and 90%), carbol xylene, and a 4:1 mixture of 50% alcohol and glacial acetic acid, then stained with an acid fuchsin solution and finally cleaned in clove oil and mounted in Canada balsam. The morphological identification was conducted through the keys proposed by Jansen and Westenberg (2015), Kozár and Konczné Benedicty (2004, 2007) and Szita et al. (2020).
Molecular characterization
The molecular characterization of the sample was carried out by sequencing the ≈ 800 COI gene part suggested by Jansen and Westenberg (2015). DNA was isolated from a single specimen of R. multiporifera, R. maasbachi and R. hibisci using the E.Z.N.A.® Tissue DNA Kit (Omega Bio-tek, Inc., Norcross, GA, USA). Non-destructive DNA extraction protocol was followed to allow morphological identification of the processed specimens. Primer pairs were PcoF1 5’-CCTTCAACTAATCATAAAAATATYAG-3’ and LepR1 5’-TAAACTTCTGGATGTCCAAAAAATCA-3’ as described in Jansen and Westenberg (2015). PCR was done according to the protocol described in Ricupero et al. (2021). Briefly, each reaction was performed in 20 µL volume with 0.85X of FailSafeTM PCR 2X PreMix F (Lucigen Corporation, Middleton, WI, USA), 0.5 µM of each primer, 1.5 units of Taq DNA Polymerase (Invitrogen, Thermo Fisher Scientific, Waltham, MA, USA) and 2 µL of DNA template. The cycling was as follows: 96 ◦C for 5 min, 35 cycles at 96 ◦C for 45 s, 45 ◦C for 1 min, 72 ◦C for 1 min, followed by a final cycle at 72 ◦C for 10 min. Reactions and cycling conditions were carried out in Applied Biosystems™ MiniAmp™ Plus Thermal Cycler. PCR products were first checked by electrophoresis using 1% agarose gel. Unsuccessful DNA amplification was obtained for R. maasbachi repeated times; therefore, no molecular identification was achieved for this species. Successful PCR products were shipped to BMR Genomics sequencing service (Padova, Italy) that purified and sequenced the amplification products through Sanger’s method. The resulting coding regions were checked for errors and trimmed for low quality in Unipro UGENE version 1.26.1. FASTA files were thus aligned to reference sequences from the National Center for Biotechnology Information (NCBI) GenBank® through Basic Local Alignment Search Tool (BLAST) sequence analysis tool for the species identification. All sequences were deposited in GenBank under accession numbers: OQ833548.1 for R. hibisci and OQ833547.1 for R. multiporifera.
Phylogenetic analysis
Evolutionary relationships among Ripersiella spp. isolates were estimated by constructing phylogenetic trees based on mtCOI sequences derived from our samples and those retrieved in GenBank in June 2023. Sequence records were screened out for their coverage within the region amplified by PcoF1 and LepR1 primers. Five selected nucleotide sequences were thus aligned with MUSCLE algorithm (Altschul et al., 1990) and their ends were trimmed to produce 695 bp alignments in Unipro UGENE version 1.26.1 (Edgar, 2004). We also screened translated mtCOI sequences for stop codons to exclude any possible mitochondrial pseudogenes that often occur in invertebrates. Phenacoccus solenopsis Tinsley (Hemiptera: Pseudococcidae) isolate mtCOI sequence (GenBank accession number MZ398132.1) was included in the dataset as outgroup. The evolutionary history was inferred by using the Maximum Likelihood (ML) method and Kimura 2-parameter model (Okonechnikov et al., 2012). Initial tree for the heuristic search was obtained automatically by applying Neighbor-Join and BioNJ algorithms to a matrix of pairwise distances estimated using the Maximum Composite Likelihood (MCL) approach and then selecting the topology with superior log likelihood value. This analysis involved 6 nucleotide sequences. Codon positions included were 1st + 2nd + 3rd + Noncoding. The reliability of the branches was estimated using 1000 bootstraps. Evolutionary analyses were conducted in MEGA X (Kimura, 1980; Kumar et al., 2018).
Results
Ripersiella multiporifera Jansen, 2008
Taxonomic identification
The slide-mounted females show elongated oval body about 1,1–1,3 mm long and 0,39 − 0,7 mm wide; antennae are 5 segmented, eyes absent, two circuli present (Fig. 2), with truncate-conical shape on abdominal segments II and III; multilocular disc pores present on venter and dorsum of head, distributed in rows on the segments of abdomen on dorsum, trilocular pores evenly distributed on dorsum and numerous on venter on head, thorax and abdomen; bitubular pores of two size: the smaller, each about 3.5 μm wide and 5 μm high (2/3–3/4 from those of dorsum) confined to venter in single transverse rows, mainly in middle of abdominal segments; the larger type (6–7 μm wide and 7–8 μm high) present on dorsum only, usually distributed on margins and submarginal areas and on midline. Tubular ducts absent on dorsum and venter.
Ripersiella multiporifera is very similar to R. saintpauliae (Williams) from which differs in having up to 45 multilocular disc pores in one to occasionally two rows on the posterior edge of segments and single ones on the rest of segment whilst R. saintpauliae has multilocular disc pores on thorax and sixth abdominal segments in small numbers up to ten per segment. Ripersiella multiporifera differs also from R. hibisci in having multilocular disc pores on the head and single rows on the thorax and first two abdominal segments and two circuli, whilst in R. hibisci, though 0–2 circuli can be present, multilocular disc pores are absent on the head and those on the thorax and first two abdominal segments are represented by single ones (Jansen, 2008).
Regarding the molecular characterization, once genomic DNA amplification was confirmed, the obtained sequence was subjected to BLAST searches and aligned to reference sequences from NCBI compared with publicly available data on GenBank. The resulting sequence matched an identity score of 100% and E-value = 0.0 with R. multiporifera isolates from the Netherlands (Accession number KM453216.1).
Ripersiella maasbachi (Jansen, 2003)
Taxonomic identification
The females have elongate-oval body with almost parallel sides 0.8–2.3 mm long and 0.38–1.2 mm wide; antennae are slender and 6-segmented, eyes present, one circulus present on abdominal segment III; multilocular disc pores present only on last 2–3 abdominal segments of venter and absent from dorsum; trilocular pores evenly scattered on dorsum and less numerous on venter where are scattered on thorax; bitubular pores of one size present on dorsum and venter; tubular ducts absent (Fig. 2).
The specimens collected in Sicily showed 10–11 multilocular disc pores on VII and 11–20 in segments on venter.
Ripersiella maasbachi is similar to Ripersiella emarai Jansen in the presence of eyes, antennae 6-segmented and the absence of dorsal multilocular disc pores, and is different in the presence of ventral multilocular disc pores on last abdominal segments and in the absence of oral collar tubular ducts (Jansen & Westenberg, 2015).
Ripersiella hibisci (Kawai & Takagi, 1971)
Taxonomic identification
The body of the adult female is elongated-oval, with a length range of 1.2–2.2 mm; antennae 5-segmented; circuli absent or numbering one or two on II and III abdominal segment; multilocular disc pores present on dorsum on abdominal segments II-V and on thorax, present also on venter in single to triple rows on III and posterior segments and on thorax; trilocular pores evenly distributed on dorsal and ventral surfaces; bitubular pores of two size: the larger (> 5 μm) present only on dorsum usually singly on margins of most segments, few on submarginal areas and occasionally on midline; the smaller (∼5 μm) present on dorsum on abdominal segments IV-V and on venter as far forward as segment IV (Williams, 1996).
The specimens collected in Sicily showed circulus absent (∼38% of observed specimens) or numbering one (∼62%) on III abdominal segment (Fig. 3).
Ripersiella hibisci is recognizable for the presence of large bitubular pores on dorsum and their absence on venter, the absence of multilocular disc pores from venter and the presence on III-V dorsal segments. This species is similar to R. saintpauliae (Williams, 1985) but the latter lacks small bitubular pores on dorsum and it has numerous multilocular disc pores on dorsum and venter (Williams, 1996).
Based on mtCOI, we confirmed amplification from genomic DNA, followed by direct sequencing and BLAST searches. The resulting sequence was aligned to reference sequences from NCBI compared with publicly available data on GenBank and yielded an identity score of 99.83% and E-value = 0.0 with R. hibisci isolates from the Netherlands (Accession number. KM453214.1).
Phylogeny
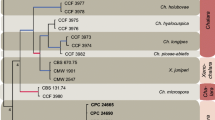
According to the phylogenetic analysis, the ML tree included two main distinct clades: one clade consisted of R. hibisci accessions, and another clade grouped accessions associated with R. multiporifera. Notably, both the clades shared a common ancestor if compared to the provided outgroup. Nevertheless, R. hibisci and R. multiporifera accessions from Italy clustered with same-species isolates coming from the Netherlands, showing no differences in their provenance (Fig. 4).
Maximum Likelihood tree, with bootstrap values, based on mtCOI sequences of Ripersiella spp. collected in Sicily (Italy) clustering with publicly available accessions from Netherlands retrieved in GenBank. Phenacoccus solenopsis (Accession n. MZ398132.1) was used as an outgroup. The tree with the highest log likelihood (-1809.31) is shown. The percentage of trees in which the associated taxa clustered together is shown next to the branches. The tree is drawn to scale, with branch lengths measured in the number of substitutions per site
Key of Rhizoecidae recorded or intercepted in Italy
For determining the species with morphological identification, a taxonomic key of adult female of Rhizoecidae reported and/or intercepted in Italy is following reported. The key was adapted from Marotta, 1992; Russo and Mazzeo, 1992; Jansen and Westenberg, 2015. According to Marotta (1995) and Russo and Mazzeo (1992) Rhizoecus mammillariae (Targioni Tozzetti, 1884), even if reported for the Italian fauna, was not included because of missing data.
-
1) Presence of bitubular or tritubular pores ……………………………….…. 2
-
Absence of bitubular and tritubular pores Ripersiella poltavae (Laing, 1929)
-
2) Presence of bitubular pores and absence of tritubular pores …………..…. 3
-
Absence of bitubular pores and presence of tritubular pores …………..…. 8
-
3) Multilocular disc pores present only on venter, with one circulus ………… 4
-
Multilocular disc pores present on venter and dorsum, with 0–1 circulus … 6
-
4) With tubular ducts; bitubular pores short or long …………………………… 5
-
Without tubular ducts; bitubular pores of one size,
-
scattered on dorsum Ripersiella maasbachi (Jansen, 2003)
-
5) Bitubular pores short, wide, 2–3 times longer than wider … Ripersiella periolana Goux, 1985
-
Bitubular pores long, narrow, 3–6 times longer than wider…. Ripersiella vidanoi Marotta & Tranfaglia, 1995
-
6) Circulus present, multilocular disc pores present on venter and dorsum,
-
scattered on head and thorax, in rows across all abdominal segments, ….…. Ripersiella lelloi Mazzeo, 1995
-
Circulus present or absents, multilocular disc pores not with this
-
combination of characters ………………………………………7
-
7) Multilocular disc pores absent on head. Small type bitubular pores 5 μm
-
wide present on venter and dorsum Ripersiella hibisci (Kawai & Takagi, 1971)
-
Multilocular disc pores present on head. Small type bitubular pores about
-
3,5 μm wide confined to venter …… Ripersiella multiporifera Jansen, 2008
-
8) Circulus present, multilocular disc pores absent ………………… 9
-
Circulus absent, multilocular disc pores present …………………10
-
9) Labium 60–70 μm long ………………… Rhizoecus albidus Goux, 1942
-
Labium 75–90 μm long ………………… Rhizoecus cacticans (Hambleton, 1946)
-
10) Antennae five-segmented ………… Rhizoecus falcifer Kunckel d’Herculais, 1878
-
Antennae six-segmented …………………………………………………………11
-
11) Tritubular pores of one size ………………………… Rhizoecus dianthi Green, 1926
-
Tritubular pores of 2–3 sizes …………………………………………………… 12
-
12) Tritubular pores of two sizes ……………………… Rhizoecus latus (Hambleton, 1946)
-
Tritubular pores of three sizes ………………… Rhizoecus americanus (Hambleton, 1946)
Discussion
We report for the first time Ripersiella multiporifera in Italy on S. trifasciata roots. This species has been described in the Netherlands where it was intercepted on Sansevieria sp. and Hoya kerrii plants originating from Thailand during phytosanitary and import inspections in commercial greenhouses (Jansen, 2008; Jansen & Westenberg, 2015). Ripersiella multiporifera is also known for China, Malaysia, Vietnam, and Indonesia (Jansen, 2008; Suh et al., 2013). Little is known about its biology and damage on host plants and, to date, it is not subject to regulatory or quarantine measures in Europe.
Ripersiella multiporifera is very similar to R. saintpauliae and R. hibisci and differences among these species were highlighted. Although R. multiporifera is difficult to discriminate from the two congeneric species because of the variability of some morphological characters (e.g., number of circuli, distribution of multilocular disc pores), the samples we collected had a combination of characters that matched with R. multiporifera according to the keys proposed by Jansen (2008) and Jansen and Westenberg (2015). The identity of the mealybugs was also corroborated through DNA amplification of the mtCOI fragment that has been suggested for species discrimination (Jansen & Westenberg, 2015).
Ripersiella maasbachi was described by Jansen in 2003 from specimens collected on bonsai trees of Segeretia thea (Osbeck) M. Johnston (Rhamnaceae) originating from China during import inspections. The species was also found on Carmona sp. (Boraginaceae), Michelia sp. (Magnoliaceae) and Serissa foetida (L. f.) Lam. (Rubiaceae) and regularly intercepted in Netherlands and England (Jansen, 2003). We collected and identified this species in Sicily and it was the first record in Italy (EPPO, 2022a). The biology and the damage it causes to host plants are not well known, and this species is not subject to regulatory or quarantine measures in Europe.
Ripersiella hibisci was described by Kawai and Takagi in 1971 (as Rhizoecus hibisci) from specimens collected on roots of cultivated potted plants in greenhouses and currently is known on 39 genera belonging to 23 families of host plants (García Morales et al., 2016). It is a polyphagous species that feeds on plant roots and often detected on potted ornamentals. First detected in Japan, it likely occurs elsewhere in South-east and East Asia and was introduced into USA and Puerto Rico (EFSA PLH PANEL, 2020). In Europe R. hibisci is not established according to EPPO (2022b) although it has been intercepted repeatedly during import inspections in Netherlands, UK and Italy where actions are taken to eradicate infestations (EFSA PLH PANEL, 2020). Although it is considered a greenhouse pest it also occurs outside where climatic conditions allow such as Japan and China and Europe also (EFSA PLH PANEL, 2020). Ripersiella hibisci is listed in Annex II of Commission Implementing Regulation (EU) 2019/2072 and regulated as a quarantine pest as a consequence of the economic and environmental impact it may cause in case of severe infestations on host plants (EFSA PLH PANEL, 2020).
Unveiling the introduction pathways of alien species is of paramount importance for assessing dispersal patterns and establishing efficient control measures (Estoup & Guillemaud, 2010). According to our phylogenetic analyses using mtCOI, data highlighted that Ripersiella spp. collected in Italy were the same of those retrieved in Netherlands (Jansen & Westenberg, 2015). This finding suggests that the origin between the species is similar. However, further studies including samples from different regions and/or different types of molecular markers are warranted to reveal the invasion route of the considered ground mealybugs (Behura, 2006).
Control measures
Although root-feeding mealybugs live in the ground and show limited ability to spread, measures to control their infestations are needed. The isolation of growing plants in association with the cleaning of tools and machinery, and the mechanical cleaning of roots are suggested to reduce the likelihood to transport mealybug eggs and post-embryonic stadia. Hot water treatments have also been used to cause adult mortality (Hu et al., 1996; Hara, 2013). Hata et al. (1996) reported that imidacloprid significantly reduced the population of R. hibisci in potted plants and Cabral and Hara (2015) reported that spirotetramat, applied as a soil drench, was able to reduce the number of R. hibisci on roots of Pisonia brunoniana. However, since repeated applications of conventional insecticides often lead to resistance outbreaks (Shankarganesh et al., 2022), further sustainable control measures can be investigated, such as the use of plant essential oils (e.g., Allium sativum) as promising results have been obtained on other invasive insect pests (Ricupero et al., 2022; Tortorici et al., 2022).
Conclusion
Among the invasive species, root mealybugs can cause a series of negative implications for the sustainability of cultivation systems because of their complex biology and the difficulties in identification. For these reasons, careful identification tools are necessary to quickly detect them and provide effective preventive measures. We reported for the first time R. multiporifera in Italy with the presence of R. maasbachi and R. hibisci through the use of morphological and molecular identification tools. A taxonomic identification key was also provided to help specialists to find the identity of Rhizoecidae species currently present in Italy. Nevertheless, continuous updates on the morphological taxonomic keys associated with new molecular tools for fast and precise identification of root mealybugs are recommended.
References
Altschul, S. F., Gish, W., Miller, W., Myers, E. W., & Lipman, D. J. (1990). Basic local alignment search tool. Journal of Molecular Biology, 215(3), 403–410.
Behura, S. K. (2006). Molecular marker systems in insects: Current trends and future avenues. Molecular Ecology, 15(11), 3087–3113.
Cabral, S. K., & Hara, A. H. (2015). Efficacy of insecticides against root mealybugs in Pisonia brunoniana, 2012. Arthropod Management Tests, 2015, 1–2. https://doi.org/10.1093/amt/tsv178
Edgar, R. C. (2004). MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Research, 32(5), 1792–1797.
EFSA PLH Panel (EFSA Panel on Plant Health). (2020). Bragard, C., Dehnen-Schmutz, K., Di Serio, F., Gonthier, P., Jacques, M. A., Jaques Miret, J. A., Justesen, A. F., Magnusson, C. S., Milonas, P., Navas-Cortes, J. A., Parnell, S., Potting, R., Reignault, P. L., Thulke, H. H., Van der Werf, W., Civera, A. V., Yuen, J., Zappalà, L., … MacLeod, A. 2020. Scientific opinion on the pest categorisation of Ripersiella hibisci. EFSA Journal, 18(6), 6178, 28 pp. https://doi.org/10.2903/j.efsa.2020.6178
EPPO. (2022a). First report of Ripersiella maasbachi in Sicilia (Italy). EPPO Reporting Service no. 6 -2022, 2022/127. Available on line at: https://gd.eppo.int/reporting/article-7358
EPPO (2022b). Eradication of Ripersiella hibisci in Italy. EPPO Reporting Service no. 01–2022, 2022/007. Available online at: https://gd.eppo.int/reporting/article-7237
Estoup, A., & Guillemaud, T. (2010). Reconstructing routes of invasion using genetic data: Why, how and so what? Molecular Ecology, 19(19), 4113–4130.
García Morales, M., Denno, B. D., Miller, D. R., Miller, G. L., Ben-Dov, Y., & Hardy, N. B. (2016). ScaleNet: A literature-based model of scale insect biology and systematics. Database. https://doi.org/10.1093/database/bav118, https://scalenet.info. Accessed on 10 Jan 2022.
Hara, A. H. (2013). Heat as a sustainable postharvest disinfestation treatment for export horticultural crops. International conference on postharvest pest and disease management in exporting horticultural crops. Acta Horticulturae, 973, 45–53.
Hata, T. Y., Hara, A. H., & Hu, B. K. S. (1996). Use of a systemic insecticide granule against root mealybugs, Hawaii, 1995, Arthropod Management Tests, 21(1), 382. https://doi.org/10.1093/amt/21.1.382
Hodgson, C. J. (2012). Comparison of the morphology of the adult males of the rhizoecine, phenacoccine and pseudococcine mealybugs (Hemiptera: Sternorrhyncha: Coccoidea), with the recognition of the family Rhizoecidae Williams. Zootaxa, 3291, 1–79.
Hu, B. K. S., Hara, A. H. & Hata, T. Y. (1996). Hot water as a potential treatment against root mealybugs, Hawaii, 1995. Arthropod Management Tests, 21, 382–383. 5I. In Burditt AK Jr (ed.). Entomological Society of America. Lanham, MD. 462 pp.
Inghilesi, A. F., Mazza, G., Cervo, R., Gherardi, F., Sposimo, P., Tricarico, E., & Zapparoli, M. (2013). Alien insects in Italy: Comparing patterns from the regional to european level. Journal of Insect Science, 13, 73. Available online: http://www.insectscience.org/13.73. Accessed 20 April 2023.
Jansen, M. G. M. (2008). A new species of the genus Ripersiella Tinsley (Hemiptera: Coccoidea: Pseudococcidae) from import interceptions in The Netherlands. Proceedings of the XI International Symposium on Scale Insect Studies, Oeiras, Portugal, 24–27 September 2007. ISA Press Lisbon, Portugal 322 pp.
Jansen, M. G. M. (2003). A new species of Rhizoecus Kunkel d’Herculais (Hemiptera, Coccoidea, Pseudococcidae) on bonsai trees. Tijdschrift voor Entomologie, 146, 297–300.
Jansen, M., & Westenberg, M. (2015). Morphological and molecular studies of a new species of the root mealybug genus Ripersiella Tinsley (Hemiptera: Coccoidea: Rhizoecidae) from greenhouses in the Netherlands and a first incursion of the american root mealybug Rhizoecus keysensis Hambleton in Europe. Tijdschrift voor Entomologie, 158, 1–19. https://doi.org/10.1163/22119434-15812049
Jucker, C., & Lupi, D. (2011). Exotic Insects in Italy: An overview on their environmental impact – In: J. L. Pujol (Ed.), The importance of biological interactions in the study of biodiversity. InTech, Available from: http://www.intechopen.com/books. Accessed 20 April 2023.
Kawai, S., & Takagi, K. (1971). Descriptions of three economically important species of root-feeding mealybugs in Japan (Homoptera: Pseudococcidae). Applied Entomology and Zoology, 6, 175–182. Tokyo.
Kaydan, M. B., Konczné Benedicty, Z., Kondo, T., Ramos-Portilla, A. A., & Szita, É. (2018). The genus Coccidella Hambleton (Hemiptera: Rhizoecidae) with description of two new species. Neotropical Entomology, 47, 369–379.
Kaydan, M. B., Konczné Benedicty, Z., Kondo, T., Ramos-Portilla, A. A., & Szita, É. (2019). Investigations on the Genus Rhizoecus (Hemiptera: Rhizoecidae) with description of two new species from South America. Neotropical Entomology, 2019(48), 809–821. https://doi.org/10.1007/s13744-019-00681-w
Kimura, M. (1980). A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. Journal of Molecular Evolution, 16, 111–120.
Kozár, F., & Konczné Benedicty, Z. (2007). Rhizoecinae of the world (p. 617). Plant Protection Institute, Hungarian Academy of Sciences Budapest.
Kozár, F., & Konczné Benedicty, Z. (2004). New species and a key of the species of the Ripersiella genus (Homoptera, Coccoidea, Pseudococcidae, Rhizoecini), with zoogeographic and phylogenetic considerations. Bollettino di Zoologia Agraria e di Bachicoltura (Milano), 36(3), 303–334.
Kumar, S., Stecher, G., Li, M., Knyaz, C., & Tamura, K. (2018). MEGA X: Molecular evolutionary genetics analysis across computing platforms. Molecular Biology and Evolution, 35(6), 1547.
Marotta, S. (1992). Ricerche su pseudococcidi (Homoptera: Coccoidea) dell’Italia centro-meridionale. Bollettino del Laboratorio di Entomologia Agraria ‘Filippo Silvestri’, 47, 63–111.
Marotta, S. (1995). Due Rhizoecus Kunckel d’Herculais, 1878 (Homoptera Coccoidea Pseudococcidae) nuovi per la fauna italiana. Bollettino di Zoologia Agraria e di Bachicoltura (Milano) Ser II, 27, 117–121.
Mazzeo, G., Longo, S., Pellizzari, G., Porcelli, F., Suma, P., & Russo, A. (2014). Exotic scale insects on ornamental plants in Italy: A never-ending story. Acta Zoologica Bulgarica, Suppl. 6, 2014, 55–61.
Mazzeo, G., Nucifora, S., & Longo, S. (2020). Definitive confirmation of establishment of Parasaissetia nigra (Nietner) (Hemiptera, Coccidae) in Sicily (Italy), with notes on its association with a new host, Syzygium myrtifolium Walp. Bulletin OEPP/EPPO Bulletin, 50(2), 295–298.
Okonechnikov, K., Golosova, O., Fursov, M., Ugene Team. (2012). Unipro UGENE: A unified bioinformatics toolkit. Bioinformatics, 28(8), 1166–1167.
Perrings, C., Williamson, M., Barbier, E. B., Delfino, D., Dalmazzone, S., Shogren, J., & Watkinson, A. (2002). Biological invasion risks and the public good: An economic perspective. Conservation Ecology, 6(1), 1.
Ricupero, M., Biondi, A., Cincotta, F., Condurso, C., Palmeri, V., Verzera, A., Zappalà, L., & Campolo, O. (2022). Bioactivity and physico-chemistry of garlic essential oil nanoemulsion in tomato. Entomologia Generalis, 42(6), 921–930. https://doi.org/10.1127/entomologia/2022/1553
Ricupero, M., Biondi, A., Russo, A., Zappalà, L., & Mazzeo, G. (2021). The cotton mealybug is spreading along the Mediterranean: First pest detection in italian tomatoes. Insects, 12(8), 675. https://doi.org/10.3390/insects12080675
Riegler, M. (2018). Insect threats to food security. Science, 361(6405), 846–846.
Russo, A., & Mazzeo, G. (1992). Rhizoecus americanus (Hambleton) e Pseudaulacaspis cockerelli (Cooley) (Homoptera Coccoidea) dannosi alle piante ornamentali in Italy. Bollettino di Zoologia Agraria e di Bachicoltura Ser. II, 24(2), 215–221.
Schrader, G., & Unger, J. G. (2003). Plant quarantine as a measure against invasive alien species: The framework of the International Plant Protection Convention and the plant health regulations in the European Union. Biological Invasions, 5(4), 357–364.
Shankarganesh, K., Ricupero, M., & Sabtharishi, S. (2022). Field evolved insecticide resistance in the cotton mealybug Phenacoccus solenopsis and its direct and indirect impacts on the endoparasitoid Aenasius arizonensis. Scientific Reports, 12(1), 16764.
Suh, S. J., Basaglia, M., & Cho, M. R. (2013). A report of two root mealybugs (Hemiptera: Rhizoecidae) on non-native ornamental plants in Korea. Korean Journal of Applied Entomology, 52(3), 255–259.
Szita, É., Konczné Benedicty, Z., Kondo, T., Portilla, R., & Kaydan, M. B. (2020). Studies on the genus Ripersiella Tinsley (Hemiptera: Coccomorpha: Rhizoecidae) in the neotropical region, with description of a new species. Zootaxa, 4851(3), 573–582.
Tortorici, S., Cimino, C., Ricupero, M., Musumeci, T., Biondi, A., Siscaro, G., & Zappalà, L. (2022). Nanostructured lipid carriers of essential oils as potential tools for the sustainable control of insect pests. Industrial Crops and Products, 181, 114766.
Williams, D.J. (1985). Mealybugs of the genus Rhizoecus (Hemiptera: Pseudococcidae) on African violets (Saintpaulia spp.) with a description of a new species from Thailand. Bulletin of Entomological Research 75, 621–624.
Williams, D. J. (1996). Four related species of root mealybugs of the genus Rhizoecus from east and southeast Asia of importance at quarantine inspection (Hemiptera: Coccoidea: Pseudococcidae). Journal of Natural History, 30(9), 1391–1403.
Williams, D. J., & Watson, G. W. (1988). The scale insects of the tropical South Pacific Region (p. 290). CAB International Institute of Entomology.
Zeng, J., Liu, Y., Zhang, H., Liu, J., Jiang, Y., Wyckhuys, K. A., & Wu, K. (2020). Global warming modifies long-distance migration of an agricultural insect pest. Journal of Pest Science, 93(2), 569–581.
Funding
Open access funding provided by Università degli Studi di Catania within the CRUI-CARE Agreement. Funding was provided by the University of Catania (Project Emergent Pests and Pathogens and Relative Sustainable Strategies − 5A722192113).
Author information
Authors and Affiliations
Contributions
Conceptualization: AR, GM; Methodology: GM, MR; Field sampling and photos: GC, FC; Morphological studies: GM, AR; Molecular studies: MR; Laboratory photos and figures editing: GM, MR; Writing - original draft preparation: GM, AR, MR; Writing - review and editing: all authors.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Mazzeo, G., Ricupero, M., Campo, G. et al. New records of Ripersiella (Tinsley) (Hemiptera: Coccomorpha: Rhizoecidae) species and identification key of Rhizoecidae in Italy. Phytoparasitica 51, 1047–1057 (2023). https://doi.org/10.1007/s12600-023-01097-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12600-023-01097-4