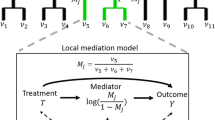
Abstract
High-dimensional mediation analysis plays an important role in recent biomedical research as a large number of mediators, such as microbiome, could modulate the effect of exposure to the outcome of interest. Most of the current studies focus on modelling independent mediators, but these methods do not consider the non-linear interactive effect between the mediators. Furthermore, it can be challenging to identify features with mediation effects from the high-dimensional mediator space. We proposed an innovative non-parametric approach to build causal mediation trees (CMT) to select important mediators and assess their non-linear interactive mediation effects on the outcome of the study. The data is recursively partitioned into subpopulations constructed by the mediators with the largest mediation effect. We aim to incorporate these non-linear interactions into the mediation framework using this approach and evaluate the total causal effect. Simulation studies were conducted to assess the performance of the CMT algorithm under different scenarios of interactive mediation effects. We applied the method to analyze vaginal microbiome sequencing data from the reproductive-age women’s study. We investigated the causal relationship between ethnic groups and the vaginal pH levels mediated by the vaginal microbiome. We identified three important microbial taxa with strong mediation effects and estimated the total effect of the mediation tree model.





Similar content being viewed by others
References
Baron RM, Kenny DA (1986) The moderator-mediator variable distinction in social psychological research: conceptual, strategic, and statistical considerations. J Personal Social Psychol 51(6):1173
Pearl J (2022) Direct and indirect effects. In: Probabilistic and Causal Inference: The Works of Judea Pearl, pp 373–392
Imai K, Keele L, Yamamoto T (2010) Identification, inference and sensitivity analysis for causal mediation effects. Stat Sci 25(1):51–71
VanderWeele TJ, Vansteelandt S (2010) Odds ratios for mediation analysis for a dichotomous outcome. Am J Epidemiol 172(12):1339–1348
VanderWeele T, Vansteelandt S (2014) Mediation analysis with multiple mediators. Epidemiol Methods 2(1):95–115
Imai K, Yamamoto T (2013) Identification and sensitivity analysis for multiple causal mechanisms: Revisiting evidence from framing experiments. Political Anal 21(2):141–171
Boca SM, Sinha R, Cross AJ, Moore SC, Sampson JN (2014) Testing multiple biological mediators simultaneously. Bioinformatics 30(2):214–220
Daniel RM, De Stavola BL, Cousens S, Vansteelandt S (2015) Causal mediation analysis with multiple mediators. Biometrics 71(1):1–14
Jérolon A, Baglietto L, Birmelé E, Alarcon F, Perduca V (2021) Causal mediation analysis in presence of multiple mediators uncausally related. Int J Biostat 17(2):191–221
Belizário JE, Faintuch J (2018) Microbiome and gut dysbiosis. In: Metabolic Interaction in Infection. Springer, pp 459–476
Wu GD, Chen J, Hoffmann C, Bittinger K, Chen Y-Y, Keilbaugh SA, Bewtra M, Knights D, Walters WA, Knight R et al (2011) Linking long-term dietary patterns with gut microbial enterotypes. Science 334(6052):105–108
Lewis JD, Chen EZ, Baldassano RN, Otley AR, Griffiths AM, Lee D, Bittinger K, Bailey A, Friedman ES, Hoffmann C et al (2015) Inflammation, antibiotics, and diet as environmental stressors of the gut microbiome in pediatric crohn’s disease. Cell Host Microbe 18(4):489–500
Kurilshikov A, Wijmenga C, Fu J, Zhernakova A (2017) Host genetics and gut microbiome: challenges and perspectives. Trends Immunol 38(9):633–647
Kim D, Zeng MY, Núñez G (2017) The interplay between host immune cells and gut microbiota in chronic inflammatory diseases. Exp Mol Med 49(5):339–339
Rothschild D, Weissbrod O, Barkan E, Kurilshikov A, Korem T, Zeevi D, Costea PI, Godneva A, Kalka IN, Bar N et al (2018) Environment dominates over host genetics in shaping human gut microbiota. Nature 555(7695):210–215
Huang Y-T, Pan W-C (2016) Hypothesis test of mediation effect in causal mediation model with high-dimensional continuous mediators. Biometrics 72(2):402–413
Zhang H, Sparks JB, Karyala SV, Settlage R, Luo XM (2015) Host adaptive immunity alters gut microbiota. The ISME J 9(3):770–781
Hakansson A, Molin G (2011) Gut microbiota and inflammation. Nutrients 3(6):637–682
Levy M, Kolodziejczyk AA, Thaiss CA, Elinav E (2017) Dysbiosis and the immune system. Nat Rev Immunol 17(4):219–232
Turnbaugh PJ, Hamady M, Yatsunenko T, Cantarel BL, Duncan A, Ley RE, Sogin ML, Jones WJ, Roe BA, Affourtit JP et al (2009) A core gut microbiome in obese and lean twins. Nature 457(7228):480–484
Qin J, Li Y, Cai Z, Li S, Zhu J, Zhang F, Liang S, Zhang W, Guan Y, Shen D et al (2012) A metagenome-wide association study of gut microbiota in type 2 diabetes. Nature 490(7418):55–60
Koeth RA, Wang Z, Levison BS, Buffa JA, Org E, Sheehy BT, Britt EB, Fu X, Wu Y, Li L et al (2013) Intestinal microbiota metabolism of l-carnitine, a nutrient in red meat, promotes atherosclerosis. Nature Med 19(5):576–585
Zhang H, Zheng Y, Zhang Z, Gao T, Joyce B, Yoon G, Zhang W, Schwartz J, Just A, Colicino E et al (2016) Estimating and testing high-dimensional mediation effects in epigenetic studies. Bioinformatics 32(20):3150–3154
Zhao Y, Luo X (2022) Pathway lasso: pathway estimation and selection with high-dimensional mediators. Stat Interface 15(1):39–50
Chén OY, Crainiceanu C, Ogburn EL, Caffo BS, Wager TD, Lindquist MA (2018) High-dimensional multivariate mediation with application to neuroimaging data. Biostatistics 19(2):121–136
Alvarez-Iglesias A, Hinde J, Ferguson J, Newell J (2017) An alternative pruning based approach to unbiased recursive partitioning. Comput Stat Data Anal 106:90–102
Morgan JN, Sonquist JA (1963) Problems in the analysis of survey data, and a proposal. J Am Stat Assoc 58(302):415–434
Athey S, Imbens G (2016) Recursive partitioning for heterogeneous causal effects. Proceed Nat Acad Sci 113(27):7353–7360
Valeri L, VanderWeele TJ (2013) Mediation analysis allowing for exposure-mediator interactions and causal interpretation: theoretical assumptions and implementation with sas and spss macros. Psychol Methods 18(2):137
Li Y, Mathur M, Yoshida K (2022) R package regmedint: extension of regression-based causal mediation analysis with effect measure modification by covariates
R Core Team (2021) R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing, Vienna, Austria. R Foundation for Statistical Computing. https://www.R-project.org/
Ravel J, Gajer P, Abdo Z, Schneider GM, Koenig SS, McCulle SL, Karlebach S, Gorle R, Russell J, Tacket CO et al (2011) Vaginal microbiome of reproductive-age women. Proceed Nat Acad Sci 108(supplement–1):4680–4687
Faust K, Sathirapongsasuti JF, Izard J, Segata N, Gevers D, Raes J, Huttenhower C (2012) Microbial co-occurrence relationships in the human microbiome. PLoS Comput Biol 8(7):1002606
Jespers V, Menten J, Smet H, Poradosú S, Abdellati S, Verhelst R, Hardy L, Buvé A, Crucitti T (2012) Quantification of bacterial species of the vaginal microbiome in different groups of women, using nucleic acid amplification tests. BMC Microbiol 12(1):1–10
Shishpal P, Patel V, Singh D, Bhor VM (2021) ph stress mediated alteration in protein composition and reduction in cytotoxic potential of gardnerella vaginalis membrane vesicles. Front Microbiol 12:723909
Serrano MG, Parikh HI, Brooks JP, Edwards DJ, Arodz TJ, Edupuganti L, Huang B, Girerd PH, Bokhari YA, Bradley SP et al (2019) Racioethnic diversity in the dynamics of the vaginal microbiome during pregnancy. Nat Med 25(6):1001–1011
Fettweis JM, Brooks JP, Serrano MG, Sheth NU, Girerd PH, Edwards DJ, Strauss JF III, Jefferson KK, Buck GA, Consortium VM et al (2014) Differences in vaginal microbiome in african american women versus women of european ancestry. Microbiology 160(Pt 10):2272
Beamer MA, Austin MN, Avolia HA, Meyn LA, Bunge KE, Hillier SL (2017) Bacterial species colonizing the vagina of healthy women are not associated with race. Anaerobe 45:40–43
Rhoades NS, Hendrickson SM, Gerken DR, Martinez K, Slayden OD, Slifka MK, Messaoudi I (2021) Longitudinal profiling of the macaque vaginal microbiome reveals similarities to diverse human vaginal communities. Msystems 6(2):01322–20
Acknowledgements
The authors would like to thank the editors and reviewers for their helpful comments and suggestions.
Funding
Wei Xu was funded by the Natural Sciences and Engineering Research Council of Canada (NSERC Grant RGPIN-2017-06672), Crohn’s and Colitis Canada (CCC Grant CCC-GEMIII), and Helmsley Charitable Trust. Yao Li was supported by NSERC Grant RGPIN-2017-06672 and CCC Grant CCC-GEMIII.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Appendices
Appendix A Estimating the Total Effect
By the definition, the Eq. (1), the NIE can be expressed as \(NIE=Y_{am_a}-Y_{a m_{a^*}}\). The estimation of \(Y_{am_a}\) are derived below:
Similarly,
.
Therefore, the average NIE can be expressed as:
The average NDE is
.
Appendix B Additional Simulations
Appendix C OTU Distribution for the Microbiome Data
See Table 9.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Li, Y., Xu, W. Causal Mediation Tree Model for Feature Identification on High-Dimensional Mediators. Stat Biosci (2023). https://doi.org/10.1007/s12561-023-09402-9
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s12561-023-09402-9