Abstract
Methylation is considered one of the proteins’ most important post-translational modifications (PTM). Plasticity and cellular dynamics are among the many traits that are regulated by methylation. Currently, methylation sites are identified using experimental approaches. However, these methods are time-consuming and expensive. With the use of computer modelling, methylation sites can be identified quickly and accurately, providing valuable information for further trial and investigation. In this study, we propose a new machine-learning model called MeSEP to predict methylation sites that incorporates both evolutionary and structural-based information. To build this model, we first extract evolutionary and structural features from the PSSM and SPD2 profiles, respectively. We then employ Extreme Gradient Boosting (XGBoost) as the classification model to predict methylation sites. To address the issue of imbalanced data and bias towards negative samples, we use the SMOTETomek-based hybrid sampling method. The MeSEP was validated on an independent test set (ITS) and 10-fold cross-validation (TCV) using lysine methylation sites. The method achieved: an accuracy of 82.9% in ITS and 84.6% in TCV; precision of 0.92 in ITS and 0.94 in TCV; area under the curve values of 0.90 in ITS and 0.92 in TCV; F1 score of 0.81 in ITS and 0.83 in TCV; and MCC of 0.67 in ITS and 0.70 in TCV. MeSEP significantly outperformed previous studies found in the literature. MeSEP as a standalone toolkit and all its source codes are publicly available at https://github.com/arafatro/MeSEP.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Post-translational modifications (PTMs) are defined as the modification of proteins after the translation process in the ribosome [1]. PTMs affect the dynamics and structure of proteins, performing a key role in numerous biological processes. There are over 200 different PTMs with various effects on the function of proteins [2]. This can be either reversible or irreversible in most cases. Covalent modifications are present in reversible reactions, while proteolytic changes can be found in irreversible reactions that move in just one way.
The methylation of lysine (K) stands as crucial among the other PTMs. Many biological activities have been demonstrated that substantially affect the lysine methylation (Kmeth) of histones. These activities included transcriptional silence or activation [3], heterochromatin compaction [4], and X-chromosome inactivation [5]. In addition, it was shown that Kmeth has a significant impact on regulating protein stability [6], subcellular localization [7], non-histone protein activity [8], and protein-protein interactions [9]. Methylation has also been shown to play an essential role in other biological interactions such as DNA repair [10], RNA processing [11], chromatin regulation [12], and signal transduction [13].
Moreover, Kmeth is implicated in several human disorders, including diabetic nephropathy, pancreatic ductal adenocarcinoma, and cancer [14,15,16], due to their function in the regulation of gene regulation. Therefore, identifying Kmeth can play a significant role in understanding different biological processes and preventing diseases. Currently, methylation in proteins is identified using experimental approaches such as mass spectrometry [17], mutagenesis of potential methylated residues [18], methylation-specific antibodies [19], and Chip-Chip [20]. These techniques are also used nowadays for identifying propionylation, glutarylation [21], malonylation [22], methylation, and a variety of lysine PTM sites [23,24,25]. Nevertheless, the time and resources needed for traditional experiments of this nature are both expensive and time-intensive. Therefore, there is a demand to develop first and cost-effective computational methods to identify Kmeth sites.
Recently, Artificial Intelligence (AI) has been played a significant role in the methodological developments for diverse problem domains, including computational biology [26, 27], cyber security [28,29,30,31], disease detection [32,33,34,35,36,37,38] and management [39,40,41,42,43,44], elderly care [45, 46], epidemiological study [47], fighting pandemic [48,49,50,51,52,53,54], healthcare [55,56,57,58,59], healthcare service delivery [60,61,62], natural language processing [63,64,65,66,67], social inclusion [68,69,70] and many more. In the last two decades, several computational techniques have been developed specifically for predicting various PTM sites. Among them, machine-learning (ML) approaches have shown promising results [5, 9, 21, 22]. The previous studies have used a variety of online protein, or PTM databases, including UniProtKB/Swiss-Prot [71, 72], dbPTM [73], and PDB [74], as well as CPLM [75] to train their models. They also used a wide range of feature encoding methods, including structural and evolutionary traits, physicochemical aspects [76,77,78,79], and sequential-based features such as PseAAC and CKSAAP [78, 80,81,82,83]. In addition, they also used different sorts of classifiers to tackle this problem. Among them, Support Vector Machine (SVM), Random Forest (RF) classifiers, and Neural Network (NN) frameworks have been widely used with promising results [82, 83, 85,86,87,88].
In an early study on PTM methylation prediction, Chen et al. [80] developed a machine-learning method called MeMo. They gathered 264 and 107 experimentally validated methylation sites for arginine and lysine, respectively, and used SVM as their employed classifier. Later on, Shien et al. proposed a technique named MASA [76]. MASA was designed to predict protein methylation sites on asparagine, arginine, glutamate, and lysine. To build this model, they used sequential and structural amino acid properties and SVM as their classification techniques. At the same time, Jianlin et al. designed the BPB-PPMS [85] model that aimed to predict lysine methylation using bi-profile Bayes feature extraction paired with SVM.
In a different study, Shi et al. introduced the PMeS [81] method, designed to enhance the prediction performance of methylation sites by using an upgraded feature encoding strategy. They utilised four sequential-based feature groups and SVM as their classification technique. They also utilised the UniProtKB [71], and Swiss-Prot [72] databases to gather 78 non-redundant proteins with 147 experimentally validated methyl lysine. In 2013, Yan et al. proposed Methcrf [77], a conditional random field (CRF) based computational predictor for identifying methylation sites in proteins for arginine and lysine residues. They utilised the MASA [76] online web-server data and coupled structural features based on Accessible Surface Area (ASA).
Later on, Qiu et al. developed a new tool called iMethyl-PseAAC [78], using pseudo amino acid composition and SVM as their classification technique. In a different study, Yinan et al. [86] proposed a new approach for predicting protein methylation sites by employing sequence conservation. Alongside this, Information Entropy (IE) was used to generate the profiles for methylated and non-methylated peptides in a broader neighbouring region around the methylation sites to create these conservation differences. In 2015, Zhe et al. developed an SVM-based method called iLM-2 L [82], which was used to predict associated methylation degrees and lysine methylation sites using the CKSAAP feature encoding approach. For relevant experimental validations, they created a training set with 226 methyllysine sites and 1518 non-methylation sites and an independent set with 14 methylation sites and 26 non-methylation sites.
In 2017, Wei et al. introduced MePred-RF [79], a new Random Forest-based model that combines enhanced feature representation capabilities and numerous discriminative sequence-based features. In a different study, Hao et al. [87] employed 3-D structural characteristics and five structural features to describe lysine methylation, including Depth Index (DPX), Electrostatic Potential (EP), Protrusion Index (CX), Residue Interaction Network (RIN), Accessible Surface Area (ASA), and Secondary Structure (SS). They used a Random Forest (RF) classifier to build the prediction model.
Recently, Sarah et al. proposed iMethylK-PseAAC [83] predictor for identifying lysine methylation sites. They made feature vectors utilising PseAAC and statistical moments and the composition of relative features. They also used an Artificial Neural Network (ANN) as their employed classifier. Most recently, Zheng et al. developed a Met-predictor [88] system that combines sequence-based data with structural attributes and employs SVM as a classifier. Lastly, Sadia et al. introduce MethEvo [84], a new machine-learning methodology designed for predicting methylation sites within proteins. MethEvo employs an evolutionary-based bi-gram profile approach for feature extraction and utilises SVM as the classification technique in its development.
Despite considerable efforts, the accuracy of predicting protein methylation sites remains limited. There are still shortcomings that need to be addressed to overcome the inadequacies of current methods in predicting lysine methylation (Kmeth) sites. In this study, we propose a novel machine-learning tool called MeSEP for methylation prediction efficiently by utilising evolutionary and structural-based information that is obtained from the Position-Specific Scoring Matrix (PSSM) and predicted local structure of proteins using SPIDER2 (SPD2) profiles. The primary samples were collected from the Protein Lysine Modification Database (PLMD) [89], which is an up-to-date data source of protein lysine modifications that have never been utilised in the literature for Kmeth site prediction. In addition, to address the imbalance issue in our dataset, we utilise the SMOTETomek hybrid method. Finally, we employed the Extreme Gradient Boost (XGBoost) classifier, which performed better than other classifiers to build our model and achieved an Accuracy (Acc) of 84.6%, Sensitivity (Sn) of 91.6%, Specificity (Sp) of 77.8%, Precision (Pre) of 0.94, the area under the curve (AUC) of 0.92, F1 score of 0.83, and Matthew’s correlation coefficient (MCC) of 0.70. To summarise, the main contribution of this study can be presented as follows:
-
Using PSSM and SPD2 profiles to represent evolutionary and structural information.
-
Using SMOTETomek-based hybrid sampling to address the imbalance issue in our training dataset and to avoid bias towards a larger class which, in this case, is the negative sample set (non-methylation sites).
-
Employing XGBoost, which outperforms different classifiers as the classification technique for methylation site prediction.
-
Outperforming previous studies by a significant margin in predicting methylation sites.
-
Building our model as a standalone toolkit which is publicly available at https://github.com/arafatro/MeSEP.
Proposed Method
In this section, we describe the dataset, the extracted evolutionary and structural features, their significance, the approach to data balancing, and the application of various classification algorithms including our base classifier. To create an effective sequence-based statistical predictor for a biological system, one must: rigorously adhere to the renowned 5-step principles of K.C. Chou [78, 82, 83, 87, 90], which are as follows: (i) the process of generating or choosing a relevant dataset for use in training and testing the predictive model, (ii) preparing biological sequence instances in an effective way that accurately reflects their fundamental relationship to the predicted target, (iii) create or build a robust algorithm that can execute the prediction process more effectively, (iv) find the right way to conduct validation tests that can be used to evaluate the performance of a predictor, and (v) implementing a useful web predictor or a standalone tool accessible to the public. Following that, our trained model was validated for accuracy using the independent test dataset. The general architecture and the entire working mechanism of MeSEP are presented in Fig. 1.
Dataset Description
The dataset utilised in this study was sourced from the Protein Lysine Modification Database [89], representing a larger and more updated version of the Compendium of Protein Lysine Modifications database [75]. The complete dataset comprises 6323 methylation sites found in 2819 protein sequences across 34 different species. For lysine and associated species information, we used the headings ‘position type’ and ‘sequence’ to select data from the methylation dataset and formed a new curated dataset. To begin, we cut the proteins into peptide sequences with a window size of 12 and extracted the unique positive and negative peptide sequences by eliminating redundant sites. We then used CD-HIT [91] to remove peptide sequences with over 40% sequential similarities across the negative dataset only. In this way, we avoid omitting the number of positive samples since they are significantly smaller than the number of negative samples. After removing redundant peptides identical to each other, the negative dataset (non-methylation sites) decreased from 127,913 to 14,027. After extracting the most relevant sequences, duplicates were removed, and the remaining unique peptide sequence data was used for training. The actual efficiency of our proposed model is evaluated using test data generated from our original data that is completely unknown to the training data. In this study, 90% of the training data and 10% of the independent test data are split at random.
Flow diagram of the entire PSI-Blast process, which applies the BLAST algorithm [98] to generate the PSSM matrix
Feature Extraction Techniques
Strings of biological sequences are the most common way to express biological data. Typically, strings are represented by one-letter notations, with each letter denoting a nucleotide or amino acid in DNA and protein, respectively. Extracting effective features to present the sequences is an important step in developing an accurate machine-learning model to predict methylation sites [79, 92,93,94]. So far, a wide range of attributes have been used to represent such sequences (e.g. proteins or peptides) [95, 96]. In this study, we incorporate features derived from both structural and evolutionary attributes. Through the use of feature extraction approaches, we can create the features based on the provided sequence in Eq. (1):
Evolutionary Based Feature
The evolutionary properties of a protein provide insights into the substitution probability of specific amino acids during the evolutionary process. It is necessary to employ more efficient ways to gather the information we are searching for. In this study, we use PSI-BLAST [97] to generate a Position-Specific Scoring Matrix (PSSM) for feature extraction. PSI-BLAST is an NCBI tool that performs numerous sequence alignments while considering mutations to find a wide range of evolutionary information. It is important to note that the database needs to be downloaded or manually constructed before running any query sequences on it. To summarise, the PSI-Blast procedure consists of the following four steps: (a) download a prepackaged database or create one using a protein dataset (b) A FASTA file (‘>’ followed by sequence data after one-line description) must be created for each protein or peptide sequence. (c) PSI-BLAST utilises a command line interface (CLI) to define alignment similarity cutoff e-value = 0.001, pseudocount = 1, and iterations = 3, and (d) The last PSI-BLAST iteration forms a PSSM matrix. Figure 2 provides an illustration of the complete procedure for ease of understanding. PSSM contains the substitution probability of a given amino acid along protein by any of the 20 amino acids depending on its position. PSSM is a matrix with the dimensions L \(\times \) 20, where L is the length of the complete protein sequence, and each column represents one of the 20 amino acids. In this study, PSSM was generated by executing three rounds of PSI-BLAST with a cutoff value of 0.001 on the non-redundant protein data bank.
Structural Based Feature
Previous studies demonstrated the effectiveness of structural features for predicting different PTMs [99,100,101,102,103]. They have also been shown to play an important role in predicting other related problems [104,105,106,107,108]. In this study, we use a predicted Secondary Structure (SS), such as Accessible Surface Area (ASA); Local Backbone Angles (which are composed of four angles called \(\phi , \psi , \theta \), and \(\tau \)), generated as the output of SPIDER2 for feature extraction. SPIDER2 is a machine-learning package that uses a deep learning framework to make predictions about the local structures of proteins. The following sections will provide insight into the particular structural properties that were explored in this study.
-
(a)
Secondary Structure (SS): The Secondary Structure of the protein represents how amino acids are folding locally in the form of helix, strands, and coil. For the Secondary Structure, the SPIDER2 tool generates dimension output for each protein, where L denotes the protein sequence length and the three columns represent the probability of amino acids to build the coil (pc), strand (pe), or helix (ph) local structures.
-
(b)
Accessible Surface Area (ASA): Within the 3D structure of a protein, ASA illustrates the approximate region that is accessible to a given solvent for each amino acid. Moreover, it reveals crucial information regarding protein structure and how it can interact with other macromolecules. In addition, it determines which amino acids are found on the surface of the protein, and that gives them a greater potential to undergo PTMs. The SPIDER2 program is used to predict an ASA value for each amino acid in a protein sequence.
-
(c)
Local Backbone Angles: Angels that are local to the backbone of proteins also serve as representations of the protein’s local structure. Torsion angles provide continuous knowledge about the local structure of proteins, whereas Secondary Structure focuses on the three discrete local structures of amino acids in a protein [103, 107, 109]. SPIDER2 predicts four local angles to present each amino acid along the protein sequence, namely \(\phi , \psi , \theta \), and \(\tau \), which are described in depth in their original works [101, 103, 110].
Formulation of Lysine Residues as Feature Vectors
In this study, transforming a protein into a peptide sequence, a lysine (methylated or non-methylated) residue is positioned in the middle with a window size of \(2n+ 1\), where n is the length of the peptide sequences upward and downward. The amino acid lysine is the residue in charge (central residue) of the methylation site, indicated as a letter notation of (K). Upward and downward must be identical in length; hence an extra residue, (X), was added at the corresponding residue ends to facilitate this. The following is the simplest definition of a peptide segment that contains all of its amino acid residues which can be written as follows:
In the context of evolutionary information, a segment E is made up of 12 amino acids that come from upward, 12 amino acids that come from downward, and a lysine residue (K) in the centre. This can be expressed as follows:
There are two sets of upward and downward amino acids for a lysine: \(R_{-\xi }\) (for \(12 \ge \sigma \ge 1\)) and \(R_{+\xi }\) (for \(1 \le \sigma \le 12\)), and then there is \(K_{i}\), the lysine residue itself, which is located in the \(i^{th}\) position of a protein sequence expressed in Eq. (3). As K is located in the middle of each 12 amino acid peptide segment in each row of the protein sequence, which is constructed using a sliding window with a length of \(2\sigma +1\); where \(\sigma \) is the total number of amino acids upward and downward.
In terms of the structural information, a segment S is made up of 3 amino acids that come from upward, 3 amino acids that come from downward, and a lysine residue in the centre that can be formulated as follows:
The mirror effect was also used to fill in the empty parts, creating a consistent window size defined by \(R_{E, S}\). This was done to ensure that there is uniformity and well-balanced.
Following the completion of the PSSM matrix overview, we then transformed the matrices into a frequency vector. This method is explained in further depth in the following. An overview of the PSSM matrix is provided below:
With a range of magnitudes in both directions, each cell in Eq. (6) may store values that are either positive or negative. Having a positive sign shows a proclivity for mutation, whilst not having one shows a lack of tendency for mutation. The magnitude of the value reflects the likelihood that the mutation will occur. According to Huang et al. [111], PSSM is portrayed in the following manners:
Only a tiny percentage of query sequences were unable to produce PSSMs; these issues were resolved by putting zeros into all of the PSSM matrix places. The size of the PSSM matrix that PSI-Blast makes is shown by the notation \(L \times n\), where L is the length of the peptide segment as a whole. The current study creates a link between the 20 amino acids and the 20 forms of alphabetical notation. Each of the 20 types of alphabetical notation points in a different direction, with L representing the length of R. The corresponding \(C_{i,j}\) amino acid central residue designates the score of the amino acid residue and is located at position \(i^{th}\) of the protein sequence, where \(i = 1, 2,..., L\), and evolves with time to become amino acid \(j=n=20\). To transpose the PSSM matrix, we created a new matrix N using the z-score normalisation of the original PSSM matrix M. The produced matrix N can be transposed using the following equation,
In Eq. (9), the formula for z-score normalisation [112] is given.
Afterwards, we utilised the following formula to calculate the average value or mean and standard deviation of 20 distinct amino acids, where \(\overline{C}_{j}\) represents the mean and \(SD(\overline{C}_{j})\) represents the standard deviation of \(\overline{C}_{j}\),
where the N matrix is symmetric, meaning that the upper and lower triangular matrices are identical. As a result of multiplying the PSSM matrix by its transpose matrix, the dimension of the resultant matrix is increased to 20 \(\text {*}\) \(L \times L\) \(\text {*}\) 20, resulting in 400-dimensional matrices. Later on, the PSSM files are utilised in the process of generating the SPD2 files.
In this study, we employed a PSSM matrix composed of three distinct components: the upper triangular matrix, the diagonal matrix, and the lower triangular matrix. Notably, this research primarily emphasises the use of the lower triangular matrix, as opposed to the upper triangular matrix. Within the realms of scientific computing and numerical analysis, a matrix is classified as sparse when it contains a substantial number of zero entries [113]. Subsequently, our focus is on the lower triangular matrix, which contains a wealth of information regarding transitions in biological evolutionary processes. As previously mentioned, the entire PSSM matrix contains a total of 400 variables. The lower triangular matrix, the upper triangular matrix (both \(190 \times 2\) = 380 elements), and the diagonal values (20 elements) combine to create the resulting feature vector. It is important to underscore that our primary emphasis is on the lower triangular matrix along with the diagonal values. This subset of the PSSM matrix comprises a total of 210 features, as 190 + 20 equals 210 features in total. Moreover, it is worth noting that the dimensions of the other SPD2 matrices generated result in \(7 \times 8\) = 56 features in total. Subsequently, we amalgamated both matrices (PSSM + SPD2) into a single 266-dimensional vector for each lysine residue. Within this 266-dimensional vector, each lysine residue is represented by a segment E and S in this 266-dimensional vector, which contains both evolutionary and structural information. In each method, normalisation coefficients are calculated to normalise the original matrices. These values of M and N are used in Eqs. (9), (10), and (11) to compute the normalised PSSM and SPD2 matrices. We decided to take this feature extraction method since its results appeared to be promising when applied to the other problem analysis [9, 92, 114,115,116].
Significance of Our Extracted Features
Here, we use the permutation model to select features that are more important and contain more discriminatory information [117]. By adjusting the value of the n_repeats parameter, it is possible to derive estimates of the relevance of features in a sample. The method divides the connection between the feature and the degree to which the feature is dependent is reflected by the degree to which the model score drops. This technique is effective since it does not rely on the model in any way, and it can be performed several times using a variety of combinations to visualise the feature’s importance.
Figure 3 illustrates the significance of features in our dataset. Some of the features have regression coefficient values that are quite close to 0. In this instance, the Extra Trees (ET) classifier is also utilised to compute the significance of features, and their Gini importance [118] has been taken into account. After determining the significance of each attribute to Gini, we assessed the importance of each Gini item and then showed the most significant features depending on their preferences. This is a typical method to eliminate one combination of features at a time to show their relative importance, which highlights the 30 most significant features utilised to build our model. If we look at the x-axis, we can visually compare two feature significance techniques for the top 30 of 266 attributes. Therefore, it is possible that accuracy does not always reflect the facts properly. Due to their improved performance, the values of Sn, MCC, F1 score, and AUC values for each of these classifiers have all increased. On the other hand, the variations in all matrices are more pronounced.
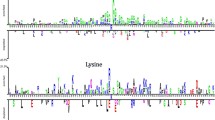
To know more about the compositional preferences of sequences close to methylation and non-methylation sites, we employed the sequence logo generator application WebLogo [119]. This tool makes it feasible to construct and show sequence profiles, as well as a visualisation of position-specific amino acid enrichment. Furthermore, it highlights the disparity between methylation (positive) peptide sequences and non-methylated sites (negatives). Figure 4a and b depict the compositional preference of amino acid frequencies around the methyl lysine and non-methyl lysine sites, respectively.
Handling Imbalanced Data
Synthetic Minority Oversampling Technique and Tomek (SMOTETomek) is a hybrid approach that is a blend of the two sampling strategies; more specifically, it combines an over-sampling strategy (SMOTE) with an under-sampling strategy (Tomek) [120]. This method combines the ability of SMOTE to make synthetic data for the minority class and the competence of Tomek to eliminate data from the majority class; the data that is closest to the minority class data samples. The working procedure of SMOTETomek is shown in Algorithm 1.
The positive sites fall into the category of the minority class, and the negative sites belong to the majority class. Note that, poor handling of imbalanced data has a terrible effect on performance as a whole [121,122,123]. Due to the considerable number of negative sites, we constructed our dataset using a technique that generates synthetic data for a small number of positive sites and excludes data samples that are too similar to minority class data samples. Therefore, we have chosen SMOTETomek because it has superior generalisation and enhanced learning capabilities when applied to previously analyzed data. Among the 26,673 (Positive: 12646 + Negative: 14027) data points, 16003 were chosen for validation and 10670 were chosen for testing; the training data reached 16550 after the SMOTETomek algorithm was applied. With an 80/20 breakdown, 80% of the data is selected at random for training, while the remaining 20% is used for testing. Following the train-test splitting, the SMOTETomek algorithm was utilised to maintain the most significant associations between the majority class and the minority class in the training dataset only. The independent test set remained unchanged and unbalanced and was not used for parameter tuning. This resulted in an imbalance ratio of 1:1 being achieved. SMOTETomek was performed on the training set to ensure that the model was generalisable. The entire procedure is presented in Table 1 for better visibility.
Classification Algorithms
The key to building an effective machine-learning approach is selecting the right classification algorithm [124,125,126]. During the model construction, we employed several classification strategies. These classifiers have been employed extensively in recent studies and have demonstrated promising results so far [21, 22, 79, 127]. We used the Support Vector Machine (SVM) [128], Multi-layer Perceptron (MLP) [129], ensemble-based Extra Trees (ET) [126], and Extreme Gradient Boosting (XGBoost) [130] classifiers, respectively. For these algorithms, default values of hyperparameters are tuned as needed in the following subsections.
Support Vector Machine
Support Vector Machine (SVM) [128] is a supervised learning method used for classification and regression analysis employing data analysis or pattern recognition. SVM can be either linear or nonlinear. It maps the x data from the input space I to a high-dimensional space H if the data are linearly inseparable–
With kernel function \(\phi (x)\)–to find the separating hyperplane. With the use of kernel function, nonlinear SVM can also exist with a nonlinear decision boundary, resulting in higher flexibility. Hyperplane dimensions are affected by the number of features. Therefore, kernel functions that can be linear, polynomial, sigmoid, radial, precomputed, or callable were utilised to cope with non-linear borders between classes. The difference between methylated and non-methylated lysine residues is calculated using a Gaussian kernel and radial basis function. To fine-tune the radial basis kernel, C = 1.0, kernel = ‘rbf’, epsilon = 0.2, and gamma = ‘scale’ were utilised.
Extra Trees
Extra Trees (ET) [126], also known as an Extremely Randomised Trees classifier, is a meta-estimator classifier that fits a diversified sub-sample of the dataset using a large number of randomised decision trees. Furthermore, averaging within an ensemble learning technique enhances predictive accuracy and reduces the overfitting issue. We utilise hyperparameters for training purposes. Consequently, we used the n_estimator parameter, and this is the actual number of trees in the forest where we used the value 10. Additionally, we used the min_sample_split=2 while developing the ET classifier.
Multilayer Perceptron
Multi-layer Perceptron (MLP) [129] is a feed-forward Artificial Neural Network. There is just one neuron in the entire network, and it applies nonlinear functions to every other neuron except the input node. MLP employs a supervised learning technique known as backpropagation for training. We used hidden layer sizes, which specify the number of neurons with a value of 100. The default function ‘relu’ is used for the activation function and returns \(f(x) = max(0, x)\) that represents the linear unit function. We set alpha = 1e-5, which is a floating point entity, and learning_rate_int = 0.001, which contains double or float entities that were used to adjust the unit size by updating their weights. Last but not least, the ‘lbfgs’ solver, which is an optimiser and a member of the family of quasi-Newton methods, and max_iter = 1000, which implies the maximum number of iterations with a value of 1000.
Extreme Gradient Boosting
Extreme Gradient Boosting (XGBoost) [130] is a decision tree-based ensemble machine-learning technique that makes use of a gradient boosting framework. To predict the problems involved with unstructured data that tends to perform better than all other frameworks or algorithms. This is why when it comes to using XGBoost, we chose to set the value of n_estimator to 300. In XGBoost, gradient boosting is utilised as a means of optimising the target. To optimise the gradient descent, an objective \(\textrm{Obj} (y, \hat{y})\) is provided, which is an iterative process that calculates as follows:
where at each iteration, to minimise the objective we enhance \(\hat{y}\) along with the direction of the gradient descent. For the objective to recall the definition, \(\textrm{Obj} = {L} + \Omega \). Afterwards, we can redefine the objective function for an iterative function given as,
Furthermore, the gradient is determined to optimise itself using gradient descent. The performance of the first and second-order gradients can also be enhanced by considering themselves.
We estimate the Second-order Taylor [131] since we lack the derivative for each objective function. Where,
By eliminating the constant terms we get,
The \(t^{th}\) step is to set an objective, and we want to reach a \(f_{t}\) goal to improve it.
Assessment of Evaluation Metrics
In this paper, we aimed to compare the effectiveness of MeSEP with that of other cutting-edge predictors when it comes to predicting methylation and non-methylated lysines using the following metrics. These seven evaluation metrics, which are extensively used in the literature as accuracy (Acc), sensitivity (Sn), specificity (Sp), precision (Pre), Area under the curve (AUC), F1 score, and Matthew’s correlation coefficient (MCC). The dataset will have +K and -K values, respectively, as indicated by Eqs. (20)–(27), and what will happen if we apply these principles to it? Consequently, any metric can be defined in the following way,
where True Positive (TP), True Negative (TN), False Positive (FP), and False Negative (FN) are shown by \(+K^{+}\), \(+K^{-}\), \(-K^{+}\), and \(-K^{-}\), respectively. TP shows how many times Methylated sites were correctly predicted. FP is the number of samples that are mistakenly labelled as Methylated when they are not. TN counts the number of non-Methylated sites that have been correctly labelled as non-Methylated. Similarly, FN is the number of methylated sites that were mistakenly labelled as non-methylated. Meanwhile, the area under the receiver-operating characteristic (ROC) curve was utilised for assessment. This curve shows how sensitivity and false positive rate (FPR) fluctuate as a function of different cut-off points in a range of values. The following is how the FPR is defined:
In light of this, the area under the ROC curve (AUROC) can be stated as follows:
where N represents the class prediction probability thresholds. It is expected that the effective predictor would have the best score in some matrices. At the very least, its sensitivity score ought to be higher when contrasted with that of other predictors [21, 87, 132]. An inability to accurately identify methylated lysine residues indicates that this method is unworthy of predicting methylation sites. In general, the quality of the predictor improves in proportion to the score that a measure achieves. Needless to say, ROC curves are used to evaluate prediction model performance where the AUC is 0 to 1. In most cases, a higher AUC suggests that the model is performing more effectively [77, 83, 132].
Results and Discussion
In this section, we present the experimental and analytical outcomes of our model and how we analysed them. The findings of each experiment were averaged across ten runs, and the mean findings were reported with all the classifiers including our base classifier and compared to previous studies as well.
Model Performance Comparison with Other Classifiers
This study aims to enhance the performance of methylation site predictions using traditional machine-learning techniques including the Support Vector Machine (SVM), Extra Trees (ET), Multi-layer Perceptron (MLP), and Extreme Gradient Boosting (XGBoost) Classifiers. We have evaluated these classifiers and separately outlined their benefits and drawbacks. In addition, we analyse the optimal ratio between training and testing data, as well as the effect of feature extraction on the output of our model MeSEP using the XGBoost classifier. In this study, we made a model based on the properties of a single residue (lysine) that incorporates the evolutionary and structural information about the residues around it. To test how well MeSEP worked, we used the testing set after the training set had been analysed by several different classifiers and the appropriate hyper-parameters had been found using 10-fold cross-validation. When it comes to identifying methylation sites, our peptide-based evolutionary PSSM and structural SPD2 feature model yield promising results. Due to the evolutionary and structural information of the neighbouring residues around each methylation lysine site, we can conclude that each methylation lysine site has its unique properties [2, 133,134,135,136].
Table 2 shows that the classifiers SVM, ET, MLP, and XGBoost were used on the training set with a value of 10 for k-fold cross-validation. This table demonstrates that the XGBoost classifier performs better than other classifiers in all measures, except Sn, which is the only metric that does not perform better. When compared with other algorithms, the XGBoost algorithm provides some improvement in terms of Acc, Sp, Precision, and MCC scores. Whereas, other classifiers show just a marginal increase over the XGBoost classifier for Sn, with a difference of less than 3%. The XGBoost and ET classifier produces the most accurate results, with an overall accuracy of 84.6%, as shown in Table 2. Compared to other classifiers, the MLP has a lower accuracy rate of 84.3%. The SVM offers us an accuracy of 83.9% in the end. Concerning accuracy scores, it is clear that the ET, MLP, and XGBoost all offer promising results that are equivalent to one another. With XGBoost on the training set, there is a 91.6% success rate for Sn (true positives), a 77.8% success rate for Sp (true negatives), and an MCC value of 0.70. When evaluated on the testing set, they provided an Sn of 91.8%, 0.67 MCC, and an Sp of 76.8%, as shown in Table 3. Furthermore, other metrics such as Acc, Sn, F1 score, and MCC are similarly high for both SVM and XGBoost classifiers, as are Sp, Precision, and AUC scores for both MLP and XGBoost classifiers. Overall, ET classifiers perform badly in terms of Acc, Sp, Precision, AUC score, and MCC across all analyses.
In addition, the ROC curve generates AUC values that are quite promising for each classification method. The scores of the 10-fold cross-validation are reflected on the ROC curve on the left, while the scores of the independent tests are reflected on the one on the right in Fig. 5.
This graphical curve illustrates that our base classifier model, MeSEP outperforms others and demonstrates the generalisability and efficacy of our approach. Note that, other classifiers depicted on the curve utilise the same extracted features as well. Their efficacy in methylation site prediction tasks is demonstrated by promising and competitive results. When we look at the left-sided figure, we can see that the AUC score for our model during cross-validation is 0.917, and when we look at the right-sided figure, we can see that the AUC score during independent testing is 0.899.
We also produced ROC (Receiver operating characteristic) curves for each of the folds used in the training set, which are shown in Fig. 6. The AUC values for the ROC curves of the training set for each fold are presented separately including (a) SVM, (b) ET, (c) MLP, and (d) XGBoost classification algorithms. These values are utilised in the validation set for the calculation of the mean. In addition, Fig. 7 demonstrates the ROC curves for the 10-fold CV vs independent test set, which have respective AUC values of 0.941 and 0.899 using our base model, XGBoost.
The research suggests that the highest AUC score for the ROC curve does not always correspond to the optimal average precision (AP) for the precision-recall (PR) curve [137]. Consequently, we assess the precision-recall curves of the classifiers employed in comparison to those obtained using our technique. This curve depicts recall and precision levels, with recall plotted along the x-axis and precision along the y-axis. Figure 8 indicates that our method yielded a PR curve AP value of 0.922. This score is equivalent to those stated for the ROC curve and represents a substantial improvement over those obtained for other classifiers. Comparing the outcomes of our proposed technique to those of other classifiers shows that the PR score is the top pick for this task. Moreover, promising results for all classifiers evaluated in this work show the usefulness of our evolutionary and structural-based properties to predict the lysine methylation sites.
Performance Comparison of Different Feature Extraction Methods
In this section, we will analyse the performance of the features that we have extracted for the model implementation. Table 4 displays the findings of the comparison and the results of utilising evolutionary and structural information together are much better than the results of using either evolutionary information or structural information individually. It is essential to keep in mind that sensitivity is the most essential parameter, seeing as how high sensitivity indicates that the predictor was able to locate methylation sites accurately [87, 132]. This highlights the importance of increasing sensitivity as the intended objective.
The fact that our model achieves more promising outcomes than other models, as can be seen in Table 5 that were just shown, is evidence of the MeSEP’s broad application and its high level of efficiency. The other classifiers are shown in the curve (see in Fig. 5) and make use of the same characteristics that were gathered from the whole data. This can be seen in the bottom right corner of the curve. Several classifiers have demonstrated good and competitive performance in methylation site prediction issues by making use of the features that we have retrieved. This demonstrates that our methodology is effective in its capacity to extract evolutionary and structural information altogether from PSSM and SPD2 profiles to tackle this challenge.
Comparison Analysis
We provide the most promising findings of MeSEP in comparison to five state-of-the-art (SOTA) predictors, named MASA [76], Methcrf [77], iMethyl-PseAAC [78], MePred-RF [79], and Met-predictor [88] in Table 5. These five predictors are regarded as the most contemporary and accurate web-based methods for predicting methylation sites. According to the results of our research, these existing predictors appear to make use of comparable classifiers. Aside from this, the datasets that were utilised by the earlier researchers were mostly out of date, as well as imbalanced, and the methods that were used to build the features could not adequately extract significant information, which led to a low rate of accuracy. From Tables 2 and 3, we can see all of the result reports are mostly focused on Acc, Sn, Sp, and MCC. Nonetheless, some other metrics provide better insights and allow us to make decisions regarding prediction performance as well. The results from the table demonstrate that our model, MeSEP, performs better than the other tools in terms of all of the criteria, except Sn and Sp rates. For Acc and MCC, the MeSEP technique obtains up to 3.79% and 0.10 improvements over MASA and iMethyl-PseAAC approach, while for AUC Score and Precision, it achieves up to 15.4% and 0.11 improvements over Methcrf and MASA.
In addition, Precision, AUC Score, and F1 score are also included during the performance assessment in our study, which has not been fully considered in previous works. When compared to prior studies in the literature, our findings show that MeSEP and the effect of our proposed features can improve lysine methylation site prediction in some evaluation matrices, such as Acc, Precision, AUC Score, and MCC. Regardless of the size of the window being used, the feature size will remain relatively the same whenever an evolutionary and structural method of feature extraction is utilised. As a consequence of this, not only does our model give better performance concerning the various evaluation scores, but it also processes an extremely large number of protein sequences that have characteristics of a constant size. This helps our technique in acquiring information that allows for the identification of patterns, and it also helps to keep the computing cost low for large volumes of data. Therefore, we also need to make our model available to the general public to contribute to the Kmeth site prediction challenge [22, 138, 139].
Note that, the State-of-the-art web servers were pre-trained with some of the protein PTM sequences used in this study for performance assessment as an independent test set. They utilised all training data and 10-fold cross-validation to test their model. Hence, it is possible that the outcomes obtained on the independent test set, which was extracted from the entire dataset, may have been subject to overestimation. The findings from those studies on the independent test set are greater than predicted. Despite this, our technique outperformed the overestimated findings. Our model is publicly accessible as a standalone application that can be downloaded and used for further improvement. The program is quite simple to use. To begin, the user must prepare the PSSM and SPD2 profile, which can be made with PSI-BLAST [97] and SPIDER2 [109] tools. The trained model is also accessible through the repository on GitHub. All of our programs, from feature extraction through model prediction, are accessible at https://github.com/arafatro/MeSEP to ease the reproducibility of our work. More specifically, Python 3.8, Scikit Learn 1.1.2, and TensorFlow 2.9.0 were utilised to build our model. In addition, we utilised PSI-BLAST 2.10.0 and SPIDER 2.0 to generate PSSM and SPD2, respectively. Finally, we used WebLogo 2.8.2 to generate the sequence logo.
Conclusion
In this study, we introduce a novel approach to the prediction of lysine methylation sites. To classify, we relied on both evolutionary and structural data that was retrieved from PSSM and SPD2 profiles, respectively. We also utilised permutation and Gini importance to determine the most significant features. Extreme Gradient Boosting (XGBoost) was used to create the MeSEP prediction model and performance on the datasets was noticeably improved. The final step in resolving all of these issues is to tune the optimal parameter for the models. In this regard, we have encountered the challenges posed by the lack of sufficient metrics for the training and test phases of model comparison.
However, there are still several issues to be concerned about, including the variance that exists across various PTMs. For this reason, it might be preferable to have high specificity as well to prevent predicting protein PTM sites. Therefore, we plan to upgrade the prediction algorithm in the future by adding an enriched dataset as the number of negative samples in any given biological dataset is almost always larger than the number of positive ones. In addition, future findings will be more precise if we can increase the number of features by adding other feature methods. To further enhance the model outcome, we will implement a hybrid voting or ensemble learning strategy. These are the actions that we plan to extend soon to improve the overall quality of our work, and we aim to implement such things promptly. The insights that were gathered from this study are not only applicable to our model but they might be also applied to the research being done on other PTM sites.
Data Availability
The code and the dataset are available at https://github.com/arafatro/MeSEP.
References
Ramazi S, Allahverdi A, Zahiri J. Evaluation of post-translational modifications in histone proteins: a review on histone modification defects in developmental and neurological disorders. J Biosci. 2020;45(1):1–29.
Beltrao P, Bork P, Krogan NJ, van Noort V. Evolution and functional cross-talk of protein post-translational modifications. Mol Syst Biol. 2013;9(1):714.
Lee DY, Teyssier C, Strahl BD, Stallcup MR. Role of protein methylation in regulation of transcription. Endocr Rev. 2005;26(2):147–70.
Grewal SI, Rice JC. Regulation of heterochromatin by histone methylation and small RNAs. Curr Opin Cell Biol. 2004;16(3):230–8.
Zhang J, Zhao X, Sun P, Ma Z. PSNO: predicting cysteine S-nitrosylation sites by incorporating various sequence-derived features into the general form of Chou’s PseAAC. Int J Mol Sci. 2014;15(7):11204–19.
Millar AH, Heazlewood JL, Giglione C, Holdsworth MJ, Bachmair A, Schulze WX. The scope, functions, and dynamics of posttranslational protein modifications. Annu Rev Plant Biol. 2019;70:119–51.
Eisenhaber B, Eisenhaber F. Posttranslational modifications and subcellular localization signals: indicators of sequence regions without inherent 3D structure? Curr Protein Pept Sci. 2007;8(2):197–203.
Hart-Smith G, Chia SZ, Low JK, McKay MJ, Molloy MP, Wilkins MR. Stoichiometry of Saccharomyces cerevisiae lysine methylation: insights into non-histone protein lysine methyltransferase activity. J Proteome Res. 2014;13(3):1744–56.
Chen C, Zhang Q, Ma Q, Yu B. LightGBM-PPI: predicting protein-protein interactions through LightGBM with multi-information fusion. Chemom Intell Lab Syst. 2019;191:54–64.
Sanford EJ, Smolka MB. A field guide to the proteomics of post-translational modifications in DNA repair. Proteomics. 2022;22(15–16):2200064.
Ruta V, Pagliarini V, Sette C. Coordination of RNA processing regulation by signal transduction pathways. Biomolecules. 2021;11(10):1475.
Tropberger P, Schneider R. Scratching the (lateral) surface of chromatin regulation by histone modifications. Nat Struct Mol Biol. 2013;20(6):657–61.
Rahimi N, Costello CE. Emerging roles of post-translational modifications in signal transduction and angiogenesis. Proteomics. 2015;15(2–3):300–9.
Sun Gd, Cui Wp, Guo Qy, Miao Ln. Histone lysine methylation in diabetic nephropathy. J Diabetes Res. 2014;2014.
Varier RA, Timmers HM. Histone lysine methylation and demethylation pathways in cancer. Biochimica et Biophysica Acta (BBA)-Reviews on Cancer. 2011;1815(1):75-89.
Roth GS, Casanova AG, Lemonnier N, Reynoird N. Lysine methylation signaling in pancreatic cancer. Curr Opin Oncol. 2018;30(1):30–7.
Afjehi-Sadat L, Garcia BA. Comprehending dynamic protein methylation with mass spectrometry. Curr Opin Chem Biol. 2013;17(1):12–9.
Qin Y, Zheng Z, Chu B, Kong Q, Ke M, Voss C, et al. Generic plug-and-play strategy for high-throughput analysis of PTM-mediated protein complexes. Anal Chem. 2022.
Ma M, Zhao X, Chen S, Zhao Y, Yang L, Feng Y, et al. Strategy based on deglycosylation, multiprotease, and hydrophilic interaction chromatography for large-scale profiling of protein methylation. Anal Chem. 2017;89(23):12909–17.
Johnson DS, Li W, Gordon DB, Bhattacharjee A, Curry B, Ghosh J, et al. Systematic evaluation of variability in ChIP-chip experiments using predefined DNA targets. Genome Res. 2008;18(3):393–403.
Shovan S, Hasan MAM, Islam MR. Improved prediction of glutarylation PTM site using evolutionary features with LightGBM resolving data imbalance issue. In: 2021 International Conference on Information and Communication Technology for Sustainable Development (ICICT4SD). IEEE; 2021. p. 141-5.
Ahmad MW, Arafat ME, Taherzadeh G, Sharma A, Dipta SR, Dehzangi A, et al. Mal-light: enhancing lysine malonylation sites prediction problem using evolutionary-based features. IEEE Access. 2020;8:77888–902.
Ao C, Jin S, Lin Y, Zou Q. Review of progress in predicting protein methylation sites. Curr Org Chem. 2019;23(15):1663–70.
Egorova K, Olenkina O, Olenina L. Lysine methylation of nonhistone proteins is a way to regulate their stability and function. Biochem Mosc. 2010;75(5):535–48.
Chen Z, Liu X, Li F, Li C, Marquez-Lago T, Leier A, et al. Large-scale comparative assessment of computational predictors for lysine post-translational modification sites. Brief Bioinform. 2019;20(6):2267–90.
Rahman MA. Gaussian process in computational biology: covariance functions for transcriptomics [phd]. University of Sheffield; 2018. Available from: https://etheses.whiterose.ac.uk/19460/.
Rakib AB, Rumky EA, Ashraf AJ, Hillas MM, Rahman MA. Mental healthcare chatbot using sequence-to-sequence learning and BiLSTM. In: Mahmud M, Kaiser MS, Vassanelli S, Dai Q, Zhong N, editors. Brain Informatics. Cham: Springer International Publishing; 2021. p. 378–87.
Islam N, et al. Towards machine learning based intrusion detection in IoT networks. Comput Mater Contin. 2021;69(2):1801–21.
Farhin F, Kaiser MS, Mahmud M. Secured smart healthcare system: blockchain and Bayesian inference based approach. In: Proc. TCCE; 2021. p. 455-65.
Ahmed S, et al. Artificial intelligence and machine learning for ensuring security in smart cities. In: Data-driven mining, learning and analytics for secured smart cities. Springer; 2021. p. 23-47.
Zaman S, et al. Security threats and artificial intelligence based countermeasures for internet of things networks: a comprehensive survey. IEEE Access. 2021;9:94668–90.
Noor MBT, Zenia NZ, Kaiser MS, Mamun SA, Mahmud M. Application of deep learning in detecting neurological disorders from magnetic resonance images: a survey on the detection of Alzheimer’s disease. Parkinson’s disease and schizophrenia Brain Inform. 2020;7(1):1–21.
Ghosh T, Al Banna MH, Rahman MS, Kaiser MS, Mahmud M, Hosen AS, et al. Artificial intelligence and internet of things in screening and management of autism spectrum disorder. Sustain Cities Soc. 2021;74: 103189.
Biswas M, Kaiser MS, Mahmud M, Al Mamun S, Hossain M, Rahman MA, et al. An XAI based autism detection: the context behind the detection. In: Proc. Brain Informatics; 2021. p. 448-59.
Wadhera T, Mahmud M. Computing hierarchical complexity of the brain from electroencephalogram signals: a graph convolutional network-based approach. In: Proc. IJCNN; 2022. p. 1-6.
Wadhera T, Mahmud M. Influences of social learning in individual perception and decision making in people with autism: a computational approach. In: Proc Brain Inform; 2022. p. 50-61.
Wadhera T, Mahmud M. Brain networks in autism spectrum disorder, epilepsy and their relationship: a machine learning approach. In: Artificial Intelligence in Healthcare: Recent Applications and Developments. Springer; 2022. p. 125-42.
Wadhera T, Mahmud M. Brain functional network topology in autism spectrum disorder: a novel weighted hierarchical complexity metric for electroencephalogram. IEEE J Biomed Health Inform. 2023:1-8.
Sumi AI, et al. fASSERT: a fuzzy assistive system for children with autism using internet of things. In: Proc. Brain Inform.; 2018. p. 403-12.
Akhund NU, et al. ADEPTNESS: Alzheimer’s disease patient management system using pervasive sensors-early prototype and preliminary results. In: Proc. Brain Inform.; 2018. p. 413-22.
Al Banna M, Ghosh T, Taher KA, Kaiser MS, Mahmud M, et al. A monitoring system for patients of autism spectrum disorder using artificial intelligence. In: Proc. Brain Informatics; 2020. p. 251-62.
Jesmin S, Kaiser MS, Mahmud M. Artificial and internet of healthcare things based Alzheimer care during COVID 19. In: Proc. Brain Inform.; 2020. p. 263-74.
Ahmed S, Hossain M, Nur SB, Shamim Kaiser M, Mahmud M, et al. Toward machine learning-based psychological assessment of autism spectrum disorders in school and community. In: Proc. TEHI; 2022. p. 139-49.
Mahmud M, Kaiser MS, Rahman MA, Wadhera T, Brown DJ, Shopland N, et al. Towards explainable and privacy-preserving artificial intelligence for personalisation in autism spectrum disorder. In: Universal Access in Human-Computer Interaction. User and Context Diversity: 16th International Conference, UAHCI 2022, Held as Part of the 24th HCI International Conference, HCII 2022, Virtual Event, June 26–July 1, 2022, Proceedings, Part II. Springer; 2022. p. 356-70.
Nahiduzzaman M, et al. Machine learning based early fall detection for elderly people with neurological disorder using multimodal data fusion. In: Proc. Brain Inform.; 2020. p. 204-14.
Biswas M, et al. Indoor navigation support system for patients with neurodegenerative diseases. In: Proc. Brain Inform.; 2021. p. 411-22.
Sadik R, Reza ML, Al Noman A, Al Mamun S, Kaiser MS, Rahman MA. COVID-19 pandemic: a comparative prediction using machine learning. International Journal of Automation, Artificial Intelligence and Machine Learning. 2020;1(1):1–16.
Mahmud M, Kaiser MS. Machine learning in fighting pandemics: a COVID-19 case study. In: COVID-19: prediction, decision-making, and its impacts. Springer; 2021. p. 77-81.
Kumar S, et al. Forecasting major impacts of COVID-19 pandemic on country-driven sectors: challenges, lessons, and future roadmap. Pers Ubiquitous Comput. 2021:1-24.
Bhapkar HR, et al. Rough sets in COVID-19 to predict symptomatic cases. In: COVID-19: Prediction, Decision-Making, and its Impacts. Springer; 2021. p. 57-68.
Satu MS, et al. Short-term prediction of COVID-19 cases using machine learning models. Appl Sci. 2021;11(9):4266.
Prakash N, et al. Deep transfer learning for COVID-19 detection and infection localization with superpixel based segmentation. Sustain Cities Soc. 2021;75: 103252.
AlArjani A, et al. Application of mathematical modeling in prediction of COVID-19 transmission dynamics. Arab J Sci Eng. 2022:1-24.
Paul A, et al. Inverted bell-curve-based ensemble of deep learning models for detection of COVID-19 from chest X-rays. Neural Comput Appl. 2022:1-15.
Mahmud M, Kaiser MS, Rahman MM, Rahman MA, Shabut A, Al-Mamun S, et al. A brain-inspired trust management model to assure security in a cloud based IoT framework for neuroscience applications. Cogn Comput. 2018;10(5):864–73.
Mahmud M, Kaiser MS, Hussain A, Vassanelli S. Applications of deep learning and reinforcement learning to biological data. IEEE Trans Neural Netw Learn Syst. 2018;29(6):2063–79.
Mahmud M, Kaiser MS, McGinnity TM, Hussain A. Deep learning in mining biological data. Cogn Comput. 2021;13(1):1–33.
Nasrin F, Ahmed NI, Rahman MA. Auditory attention state decoding for the quiet and hypothetical environment: a comparison between bLSTM and SVM. In: Kaiser MS, Bandyopadhyay A, Mahmud M, Ray K, editors. Proceedings of TCCE. Advances in Intelligent Systems and Computing. Singapore: Springer; 2021. p. 291-301.
Rahman MA, Brown DJ, Mahmud M, Shopland N, Haym N, Sumich A, et al. Biofeedback towards machine learning driven self-guided virtual reality exposure therapy based on arousal state detection from multimodal data. In: Proc. BI2022; 2022. p. 1-12.
Farhin F, Kaiser MS, Mahmud M. Towards secured service provisioning for the internet of healthcare things. In: Proc. AICT; 2020. p. 1-6.
Kaiser MS, et al. 6G access network for intelligent internet of healthcare things: opportunity, challenges, and research directions. In: Proc. TCCE; 2021. p. 317-28.
Biswas M, et al. ACCU3RATE: a mobile health application rating scale based on user reviews. PloS One. 2021;16(12).
Rabby G, et al. A flexible keyphrase extraction technique for academic literature. Procedia Comput Sci. 2018;135:553–63.
Rabby G, Azad S, Mahmud M, Zamli KZ, Rahman MM. Teket: a tree-based unsupervised keyphrase extraction technique. Cogn Comput. 2020;12(4):811–33.
Adiba FI, Islam T, Kaiser MS, Mahmud M, Rahman MA. Effect of corpora on classification of fake news using naive Bayes classifier. International Journal of Automation, Artificial Intelligence and Machine Learning. 2020 Oct;1(1):80-92. Number: 1. Available from: https://researchlakejournals.com/index.php/AAIML/article/view/45.
Das S, Yasmin MR, Arefin M, Taher KA, Uddin MN, Rahman MA. Mixed Bangla-English spoken digit classification using convolutional neural network. In: Kaiser MS, Kasabov N, Iftekharuddin K, Zhong N, editors. Mahmud M. Applied intelligence and informatics. Communications in computer and information science. Cham: Springer international publishing; 2021. p. 371–83.
Nawar A, Toma NT, Al Mamun S, Kaiser MS, Mahmud M, Rahman MA. Cross-content recommendation between movie and book using machine learning. In: 2021 IEEE 15th International Conference on Application of Information and Communication Technologies (AICT); 2021. p. 1-6.
Rahman MA, Brown DJ, Shopland N, Burton A, Mahmud M. Explainable multimodal machine learning for engagement analysis by continuous performance test. In: Stephanidis C, editor. Antona M. Universal Access in Human-Computer Interaction. User and Context Diversity. Lecture Notes in Computer Science. Cham: Springer International Publishing; 2022. p. 386–99.
Rahman MA, Brown DJ, Shopland N, Harris MC, Turabee ZB, Heym N, et al. Towards machine learning driven self-guided virtual reality exposure therapy based on arousal state detection from multimodal data. In: Mahmud M, He J, Vassanelli S, van Zundert A, Zhong N, editors., et al., Brain Informatics. Cham: Springer International Publishing; 2022. p. 195–209.
Mahmud M, Kaiser MS, Rahman MA. Towards explainable and privacy-preserving artificial intelligence for personalisation in autism spectrum disorder. In: Stephanidis C, editor. Antona M. Universal access in human-computer interaction. User and context diversity. Lecture notes in computer science. Cham: Springer international publishing; 2022. p. 356–70.
Bairoch A, Estreicher A, Boeckmann B, O’Donovan C, Gasteiger E, Phan I, et al. The SWISS-PROT protein knowledgebase and its supplement TrEMBL in 2003. Nucleic Acids Res. 2003;31:365–70.
Farriol-Mathis N, Garavelli JS, Boeckmann B, Duvaud S, Gasteiger E, Gateau A, et al. Annotation of post-translational modifications in the Swiss-Prot knowledge base. Proteomics. 2004;4(6):1537–50.
Huang KY, Su MG, Kao HJ, Hsieh YC, Jhong JH, Cheng KH, et al. dbPTM 2016: 10-year anniversary of a resource for post-translational modification of proteins. Nucleic Acids Res. 2016;44(D1):D435-46.
Burley SK, Berman HM, Kleywegt GJ, Markley JL, Nakamura H, Velankar S. Protein Data Bank (PDB): the single global macromolecular structure archive. Protein Crystallography. 2017:627-41.
Liu Z, Wang Y, Gao T, Pan Z, Cheng H, Yang Q, et al. CPLM: a database of protein lysine modifications. Nucleic Acids Res. 2014;42(D1):D531-6.
Shien DM, Lee TY, Chang WC, Hsu JBK, Horng JT, Hsu PC, et al. Incorporating structural characteristics for identification of protein methylation sites. J Comput Chem. 2009;30(9):1532–43.
Xu Y, Ding J, Huang Q, Deng NY. Prediction of protein methylation sites using conditional random field. Protein Pept Lett. 2013;20(1):71–7.
Qiu WR, Xiao X, Lin WZ, Chou KC. iMethyl-PseAAC: identification of protein methylation sites via a pseudo amino acid composition approach. BioMed Res Int. 2014;2014.
Wei L, Xing P, Shi G, Ji Z, Zou Q. Fast prediction of protein methylation sites using a sequence-based feature selection technique. IEEE/ACM Trans Comput Biol Bioinform. 2017;16(4):1264–73.
Chen H, Xue Y, Huang N, Yao X, Sun Z. MeMo: a web tool for prediction of protein methylation modifications. Nucleic Acids Res. 2006;34(suppl_2):W249-53.
Shi SP, Qiu JD, Sun XY, Suo SB, Huang SY, Liang RP. PMeS: prediction of methylation sites based on enhanced feature encoding scheme. PloS One. 2012;7(6): e38772.
Ju Z, Cao JZ, Gu H. iLM-2L: a two-level predictor for identifying protein lysine methylation sites and their methylation degrees by incorporating K-gap amino acid pairs into Chou’s general PseAAC. J Theor Biol. 2015;385:50–7.
Ilyas S, Hussain W, Ashraf A, Khan YD, Khan SA, Chou KC. iMethylK-PseAAC: improving accuracy of lysine methylation sites identification by incorporating statistical moments and position relative features into general PseAAC via Chou’s 5-steps rule. Curr Genomics. 2019;20(4):275–92.
Islam S, Mugdha SBS, Dipta SR, Arafat ME, Shatabda S, Alinejad-Rokny H, Dehzangi I. MethEvo: an accurate evolutionary information-based methylation site predictor Neural Comput Applic; 2022. p. 2749-56.
Shao J, Xu D, Tsai SN, Wang Y, Ngai SM. Computational identification of protein methylation sites through bi-profile Bayes feature extraction. PloS One. 2009;4(3): e4920.
Shi Y, Guo Y, Hu Y, Li M. Position-specific prediction of methylation sites from sequence conservation based on information theory. Sci Rep. 2015;5(1):1–14.
Qiu H, Guo Y, Yu L, Pu X, Li M. Predicting protein lysine methylation sites by incorporating single-residue structural features into Chou’s pseudo components. Chemom Intell Lab Syst. 2018;179:31–8.
Zheng W, Wuyun Q, Cheng M, Hu G, Zhang Y. Two-level protein methylation prediction using structure model-based features. Sci Rep. 2020;10(1):1–15.
Xu H, Zhou J, Lin S, Deng W, Zhang Y, Xue Y. PLMD: an updated data resource of protein lysine modifications. J Genet Genomics. 2017;44(5):243–50.
Chou KC. Some remarks on protein attribute prediction and pseudo amino acid composition. J Theor Biol. 2011;273(1):236–47.
Li W, Godzik A. Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics. 2006;22(13):1658–9.
Liu ZP, Wu LY, Wang Y, Zhang XS, Chen L. Prediction of protein-RNA binding sites by a random forest method with combined features. Bioinformatics. 2010;26(13):1616–22.
You ZH, Lei YK, Zhu L, Xia J, Wang B. Prediction of protein-protein interactions from amino acid sequences with ensemble extreme learning machines and principal component analysis. In: BMC bioinformatics. vol. 14. BioMed Central; 2013. p. 1-11.
Bonidia RP, Domingues DS, Sanches DS, de Carvalho AC. MathFeature: feature extraction package for DNA, RNA and protein sequences based on mathematical descriptors. Brief Bioinform. 2022;23(1):bbab434.
Khatun S, Hasan M, Kurata H. Efficient computational model for identification of antitubercular peptides by integrating amino acid patterns and properties. FEBS Lett. 2019;593(21):3029–39.
Zhang D, Xu ZC, Su W, Yang YH, Lv H, Yang H, et al. iCarPS: a computational tool for identifying protein carbonylation sites by novel encoded features. Bioinformatics. 2021;37(2):171–7.
Schäffer AA, Aravind L, Madden TL, Shavirin S, Spouge JL, Wolf YI, et al. Improving the accuracy of PSI-BLAST protein database searches with composition-based statistics and other refinements. Nucleic Acids Res. 2001;29(14):2994–3005.
Sansom C. Database searching with DNA and protein sequences: an introduction. Brief Bioinform. 2000;1(1):22–32.
Ahmed F, Dehzangi I, Hasan MM, Shatabda S. Accurately predicting microbial phosphorylation sites using evolutionary and structural features. Gene. 2023;851: 146993.
Dehzangi I, Sharma A, Shatabda S. iProtGly-SS: a tool to accurately predict protein glycation site using structural-based features. In: Computational Methods for Predicting Post-Translational Modification Sites. Springer; 2022. p. 125-34.
Lyons J, Dehzangi A, Heffernan R, Sharma A, Paliwal K, Sattar A, et al. Predicting backbone C\(\alpha \) angles and dihedrals from protein sequences by stacked sparse auto-encoder deep neural network. J Comput Chem. 2014;35(28):2040–6.
López Y, Dehzangi A, Lal SP, Taherzadeh G, Michaelson J, Sattar A, et al. SucStruct: prediction of succinylated lysine residues by using structural properties of amino acids. Anal Biochem. 2017;527:24–32.
Heffernan R, Dehzangi A, Lyons J, Paliwal K, Sharma A, Wang J, et al. Highly accurate sequence-based prediction of half-sphere exposures of amino acid residues in proteins. Bioinformatics. 2016;32(6):843–9.
Faraggi E, Zhang T, Yang Y, Kurgan L, Zhou Y. SPINE X: improving protein secondary structure prediction by multistep learning coupled with prediction of solvent accessible surface area and backbone torsion angles. J Comput Chem. 2012;33(3):259–67.
Chowdhury SY, Shatabda S, Dehzangi A. iDNAProt-ES: identification of DNA-binding proteins using evolutionary and structural features. Sci Rep. 2017;7(1):1–14.
Shatabda S, Saha S, Sharma A, Dehzangi A. iPHLoc-ES: identification of bacteriophage protein locations using evolutionary and structural features. J Theor Biol. 2017;435:229–37.
Dehzangi A, López Y, Lal SP, Taherzadeh G, Sattar A, Tsunoda T, et al. Improving succinylation prediction accuracy by incorporating the secondary structure via helix, strand and coil, and evolutionary information from profile bigrams. PloS One. 2018;13(2): e0191900.
Reddy HM, Sharma A, Dehzangi A, Shigemizu D, Chandra AA, Tsunoda T. GlyStruct: glycation prediction using structural properties of amino acid residues. BMC Bioinf. 2019;19(13):55–64.
Heffernan R, Paliwal K, Lyons J, Dehzangi A, Sharma A, Wang J, et al. Improving prediction of secondary structure, local backbone angles and solvent accessible surface area of proteins by iterative deep learning. Sci Rep. 2015;5(1):1–11.
Yang Y, Heffernan R, Paliwal K, Lyons J, Dehzangi A, Sharma A, et al. Spider2: a package to predict secondary structure, accessible surface area, and main-chain torsional angles by deep neural networks. In: Prediction of protein secondary structure. Springer; 2017. p. 55-63.
Huang C, Yuan J. Using radial basis function on the general form of Chou’s pseudo amino acid composition and PSSM to predict subcellular locations of proteins with both single and multiple sites. Biosystems. 2013;113(1):50–7.
Zhang Z, Gong Y, Gao B, Li H, Gao W, Zhao Y, et al. SNAREs-SAP: SNARE proteins identification with PSSM Profiles. Front Genet. 2021;12.
Buluc A, Gilbert JR. Challenges and advances in parallel sparse matrix-matrix multiplication. In: 2008 37th international conference on parallel processing. IEEE; 2008. p. 503-10.
Long H, Liao B, Xu X, Yang J. A hybrid deep learning model for predicting protein hydroxylation sites. Int J Mol Sci. 2018;19(9):2817.
Jensen LJ, Gupta R, Blom N, Devos D, Tamames J, Kesmir C, et al. Prediction of human protein function from post-translational modifications and localization features. J Molecular Biol. 2002;319(5):1257–65.
Paliwal KK, Sharma A, Lyons J, Dehzangi A. A tri-gram based feature extraction technique using linear probabilities of position specific scoring matrix for protein fold recognition. IEEE Trans Nanobioscience. 2014;13(1):44–50.
Altmann A, Toloşi L, Sander O, Lengauer T. Permutation importance: a corrected feature importance measure. Bioinformatics. 2010;26(10):1340–7.
Menze BH, Kelm BM, Masuch R, Himmelreich U, Bachert P, Petrich W, et al. A comparison of random forest and its Gini importance with standard chemometric methods for the feature selection and classification of spectral data. BMC Bioinf. 2009;10(1):1–16.
Crooks GE, Hon G, Chandonia JM, Brenner SE. WebLogo: a sequence logo generator. Genome Res. 2004;14(6):1188–90.
Ning Q, Zhao X, Ma Z. A novel method for identification of glutarylation sites combining borderline-SMOTE with Tomek links technique in imbalanced data. IEEE/ACM Trans Comput Biol Bioinform. 2021.
Prati RC, Batista GE, Monard MC. Data mining with imbalanced class distributions: concepts and methods. In: IICAI; 2009. p. 359-76.
He H, Garcia EA. Learning from imbalanced data. IEEE Trans Knowl Data Eng. 2009;21(9):1263–84.
Kumar P, Bhatnagar R, Gaur K, Bhatnagar A. Classification of imbalanced data: review of methods and applications. In: IOP conference series: Materials science and engineering. vol. 1099. IOP Publishing; 2021. p. 012077.
Modhukur V, Sharma S, Mondal M, Lawarde A, Kask K, Sharma R, et al. Machine learning approaches to classify primary and metastatic cancers using tissue of origin-based DNA methylation profiles. Cancers. 2021;13(15):3768.
Dipta SR, Taherzadeh G, Ahmad MW, Arafat ME, Shatabda S, Dehzangi A. SEMal: accurate protein malonylation site predictor using structural and evolutionary information. Comput Biol Med. 2020;125.
Arafat ME, Ahmad MW, Shovan S, Dehzangi A, Dipta SR, Hasan MAM, et al. Accurately predicting glutarylation sites using sequential bi-peptide-based evolutionary features. Genes. 2020;11(9):1023.
Ding CH, Dubchak I. Multi-class protein fold recognition using support vector machines and neural networks. Bioinformatics. 2001;17(4):349–58.
Li S, Li H, Li M, Shyr Y, Xie L, Li Y. Improved prediction of lysine acetylation by support vector machines. Protein Pept Lett. 2009;16(8):977–83.
Banerjee S, Ghosh D, Basu S, Nasipuri M. JUPred_MLP: prediction of phosphorylation sites using a consensus of MLP classifiers. In: proceedings of the 4th international conference on frontiers in intelligent computing: Theory and applications (FICTA) 2015. Springer; 2016. p. 35-42.
Yu J, Shi S, Zhang F, Chen G, Cao M. PredGly: predicting lysine glycation sites for Homo sapiens based on XGboost feature optimization. Bioinformatics. 2019;35(16):2749–56.
Zhang L, Bian W, Qu W, Tuo L, Wang Y. Time series forecast of sales volume based on XGBoost. In: J Phys Conf Ser vol. 1873. IOP Publishing; 2021. p. 012067.
Azim SM, Sharma A, Noshadi I, Shatabda S, Dehzangi I. A convolutional neural network based tool for predicting protein AMPylation sites from binary profile representation. Sci Rep. 2022;12(1):1–7.
Dehzangi A, López Y, Lal SP, Taherzadeh G, Michaelson J, Sattar A, et al. PSSM-Suc: accurately predicting succinylation using position specific scoring matrix into bigram for feature extraction. J Theor Biol. 2017;425:97–102.
Kumar P, Joy J, Pandey A, Gupta D. PRmePRed: a protein arginine methylation prediction tool. PloS One. 2017;12(8).
Martin C, Zhang Y. The diverse functions of histone lysine methylation. Nat Rev Mol Cell Biol. 2005;6(11):838–49.
Lanouette S, Mongeon V, Figeys D, Couture JF. The functional diversity of protein lysine methylation. Mol Syst Biol. 2014;10(4):724.
Davis J, Goadrich M. The relationship between Precision-Recall and ROC curves. In: Proceedings of the 23rd international conference on Machine learning; 2006. p. 233-40.
Shovan S, Hasan MAM, Islam MR. Accurate prediction of formylation PTM site using multiple feature fusion with lightgbm resolving data imbalance issue. In: 2020 23rd International Conference on Computer and Information Technology (ICCIT). IEEE; 2020. p. 1-6.
Shovan S, Ahmed B. Enhanced characterization performance of propionylation PTM utilizing multiple feature fusion. In: Proceedings of the 2nd International Conference on computing advancements; 2022. p. 1-5.
Acknowledgements
Research reported in this work was supported in part by the fellowship from the Information and Communication Technology Division (ICTD) of the Government of the People’s Republic of Bangladesh in the 2021–2022 fiscal year. The authors would like to thank the Institute of Information Technology of Jahangirnagar University for supporting this research. Mufti Mahmud is supported by the PHASE IV AI project (agreement no: 101095384) funded by the European Commission through the Horizon Europe programme.
Author information
Authors and Affiliations
Contributions
This work was carried out in close collaboration between all co-authors. MEA and MSK first conceived and initiated this study. MEA, MWA, and SMS defined the research theme and performed the experiments. MEA, MWA, SMS, MM, and MSK wrote the manuscript. TUH and NI helped with the figures and literature review. MM, MSK mentored and analytically reviewed the paper. All the authors have seen and approved the final version of the manuscript.
Corresponding authors
Ethics declarations
Ethical Approval
All procedures performed in studies involving human participants were following the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki Declaration and its later amendments or comparable ethical standards.
Informed Consent
Informed consent was obtained from all individual participants included in the study.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Arafat, M.E., Ahmad, M.W., Shovan, S.M. et al. Accurate Prediction of Lysine Methylation Sites Using Evolutionary and Structural-Based Information. Cogn Comput 16, 1300–1320 (2024). https://doi.org/10.1007/s12559-024-10268-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12559-024-10268-2