Abstract
Purpose
We generated methods for evaluating clinical outcomes including treatment response in oncology using the unstructured data from electronic health records (EHR) in Japanese language.
Methods
This retrospective analysis used medical record database and administrative data of University of Miyazaki Hospital in Japan of patients with lung/breast cancer. Treatment response (objective response [OR], stable disease [SD] or progressive disease [PD]) was adjudicated by two evaluators using clinicians’ progress notes, radiology reports and pathological reports of 15 patients with lung cancer (training data set). For assessing key terms to describe treatment response, natural language processing (NLP) rules were created from the texts identified by the evaluators and broken down by morphological analysis. The NLP rules were applied for assessing data of other 70 lung cancer and 30 breast cancer patients, who were not adjudicated, to examine if any difference in using key terms exist between these patients.
Results
A total of 2,039 records in progress notes, 131 in radiology reports and 60 in pathological reports of 15 patients, were adjudicated. Progress notes were the most common primary source data for treatment assessment (60.7%), wherein, the most common key terms with high sensitivity and specificity to describe OR were “reduction/shrink”, for SD were “(no) remarkable change/(no) aggravation)” and for PD were “(limited) effect” and “enlargement/grow”. These key terms were also found in other larger cohorts of 70 patients with lung cancer and 30 patients with breast cancer.
Conclusion
This study demonstrated that assessing response to anticancer therapy using Japanese EHRs is feasible by interpreting progress notes, radiology reports and Japanese key terms using NLP.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
Real-world data (RWD) represent data collected from diverse areas of routine clinical practice and are considered as mutually complementary to randomized controlled trials [1]. The RWD originates from electronic health records (EHRs), health insurance claims data, registries (disease and product) and pragmatic clinical trials [2]. In Japan, most RWD studies are based on administrative databases, including insurance claims and diagnosis procedure combination (DPC) data. The research evaluating the outcomes of drug/treatment is not feasible using health insurance claims data or DPC data in Japan as they are currently limited to charting review methods at limited facilities and incur high cost. On the other hand, there is a growing research interest in using electronic health record (EHR) databases for generating RWD.
EHRs, simply, are electronic clinical information systems used and maintained by healthcare systems to collect, store and present longitudinal electronic data collected during the delivery of healthcare [3,4,5]. EHR databases contain a wider range of variables recorded during medical examination compared to administrative databases. EHRs capture large arrays of progressive medical information from patients at different time points in the disease history and across different clinical care systems. EHRs contain various types of patient-level structured data, such as demographics, diagnoses, medications, vital signs and laboratory data [3]. Utilizing structured data for RWD collection is convenient; however, EHRs also contain considerable amounts of unstructured data, such as progress notes which remains a significant challenge in using EHRs for RWD collection [6]. The unstructured data may contain key patient information absent from the structured data. Indeed, most clinical outcomes are unavailable from structured data but available from unstructured data [7]. To evaluate unstructured EHR data, researchers are required to manually review a patient’s chart or to use advanced technologies, such as artificial intelligence based on natural language processing (NLP). NLP has potential to synthesize standardized text strings from unstructured notes containing unprotected health information. However, NLP methods are still evolving and are not widely accessible owing to associated high costs and the operating expertise required.
Recently, limited oncology studies in the USA have evaluated clinical outcomes, such as tumor response and disease progression from unstructured radiology reports using EHR databases [8,9,10]. We aim to utilize the developed method to evaluate comparative effectiveness of treatments using EHR databases which could generate evidence timelier than the evidence generated using the primary data collection [11, 12]. This is expected to support the decision on treatment selection in real world setting. To achieve this objective, NLP method is required to handle the routinely collected large-scale EHR data. Some studies have specifically demonstrated the use of NLP in assessing clinical outcomes in patients with cancer using EHRs [13, 14]. However, there have been challenges pertaining to use of non-English language such as word segmentation and functional expression in the Japanese language [15, 16]. We believe there is a dearth of data reporting oncology outcomes using unstructured data in Japanese language from EHRs. In this descriptive study, we aimed to generate initial methods for extracting clinical outcomes of response to anticancer drugs such as tumor response and treatment discontinuation in patients with cancer utilizing the EHR of University of Miyazaki Hospital in Japanese language. The hospital is one of the major hospitals among the ~ 106 hospitals participating in the Japanese EHR under “Millennial Medical Record Project” [17]. We also applied NLP for morphologically extracting key terms describing the treatment response in unstructured text data of Japanese language so that the NLP method allows researchers to generate evidence using a large scale EHR database.
2 Methods
2.1 Study design and patient population
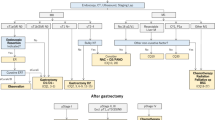
This was a retrospective study of patients with cancer using databases of medical records and administrative data of University of Miyazaki Hospital in Japan. The study included patients ≥ 18 years of age with a diagnosis of lung cancer or breast cancer who visited or were admitted to the University of Miyazaki Hospital between April 2018 and September 2020 and who had received anticancer therapy having radiology reports and progress notes during the treatment. We selected lung cancer patients with longer follow-ups for evaluating the treatment response. However, we also applied our method to breast cancer patients to conduct preliminary examination regarding any remarkable differences in using key terms between lung and breast cancer.
Patients who requested the suspension of personal information use, or those who were prescribed unapproved drugs, and those who were considered ineligible for inclusion by the principal investigator, were excluded from the study.
2.2 Study outcomes
Main study outcomes were line of treatment of anticancer drug therapy, response to treatment in terms of objective response (OR, i.e. complete or partial response), stable disease (SD) or progressive disease (PD) in real-world. We also aimed to identify key terms used to describe treatment responses in the medical records.
Criteria for evaluating response to drug treatment, including treatment effectiveness and decision for continuing/discontinuing treatment in clinical practice, were defined based on the basic principles of RECIST criteria [18]. Adjudication of treatment response was performed by two evaluators (experienced physicians) using clinicians’ progress notes, radiology reports and pathological reports according to the following criteria of response to treatment: OR – any shrinkage of tumor compared to baseline observed in radiology images; PD – any progression compared to baseline or treatment discontinuation due to insufficient efficacy or intolerance; SD – the outcome when neither OR nor SD was observed (Online Resource, Supplementary Methods). Baseline was considered to be the start date ± 1 month of each treatment line. Radiology reports included CT, MRI and PET-CT scans; whereas simple X-ray interpretation results were included in progress notes.
Among all eligible patients, 15 patients with lung cancer who had longer follow-ups were selected for the adjudication by two evaluators as a training set (Fig. 1). Two evaluators reviewed all the progress notes, radiology reports and pathological reports for the 15 patients and adjudicated treatment response and identified texts of the grounds for the adjudication. From identified texts of treatment response by the evaluators, key terms for the treatment response were extracted and broken down/selected by morphological analysis and were then used for generating the NLP rules. These NLP rules were applied to 70 patients with lung cancer who were not assessed by the evaluators and 30 patients with breast cancer, respectively.
For identifying lines of treatment, an algorithm was set because medical records do not include information regarding lines of treatment. An initial set of drugs administered at first dosing after diagnosis was regarded as first-line treatment. A set of drugs administered at the earliest next dosing that was different from those at the first dosing was regarded as the second or later line. If the same drug was used in multiple treatment lines drug treatment records of the patient were individually reviewed. Lines of treatment generated by this algorithm for patients in the training set were also confirmed by two evaluators.
2.3 Data analysis
The anonymized patient information was analyzed within the intra-net of University of Miyazaki Hospital. Drug treatments were summarized as per the lines of treatment according to the algorithm cited above. Best responses in each treatment line which were adjudicated by two evaluators were summarized and a concordance correlation was evaluated by calculating the kappa coefficient. A summary of data sources (progress notes, radiology reports and pathological reports) that contributed to the adjudication was also written. Sensitivity and specificity of key terms extracted from the text data by morphological analysis associated with each treatment response (OR, SD, PD) were calculated. The key terms for treatment response were extracted from identified texts by the evaluators and were broken down into parts of speech that was further classified into negative/positive context by morphological analysis. For the morphological analysis, Text Mining Studio of NTT DATA Mathematical Systems Inc. software was used [19] and a dictionary specialized for treatment responses in cancer patients with Japanese medical terminologies provided by Medical Information System Development Center of a General Incorporated Association [20], RECIST Guideline version 1.1 of Japanese translation and others in addition to the software’s standard dictionary was created [18, 21]. Frequency, proportion and the 95% confidence intervals of key terms were calculated for patients who were not assessed by evaluators. Missing values for each variable were summarized but were not counted for calculating summary statistics.
3 Results
3.1 Patient disposition and demographics
Between April 2018 and September 2020, of 83,894 patients with EHRs who were admitted in the University of Miyazaki hospital, 1,930 patients were diagnosed with lung cancer or breast cancer. Among eligible patients, the pre-specified sample size of 115 patients were selected as described in the methods. A total of 15 patients with lung cancer with longer follow-ups were selected for adjudication, and the other 100 patients were not assessed by evaluators (70 patients with lung cancer and 30 patients with breast cancer). Mean age of the patients was 67 years and the majority were females (67%). Since this study included patients with breast cancer, the percentage of female patients was higher in the overall cohort; however, the majority of patients with lung cancer assessed by evaluator were males (10/15, 66.7%). Most patients (64/115, 55.7%) were treated for stage 3/4 cancer and 62.6% (72/115) were hospitalized at the time of the analysis due to primary cancer. Recurrence of primary cancer was seen in 1.7% (2/115) of patients; metastasis primary cancer in 28.7% (33/115); and multiple primary cancer in 7.8% (9/115) patients (Table 1).
3.2 Pharmacotherapy for lung cancer
Of 85 patients with lung cancer, all patients (100%) received first-line and 27% patients received second-line therapy according to the algorithm (Fig. 2a). Among the patients with breast cancer, 100% patients received first-line therapy and 60% received second-line therapy. We could identify a single drug or multiple drug combinations for each treatment line. Carboplatin + paclitaxel, pembrolizumab + carboplatin + pemetrexed sodium, cisplatin + docetaxel, and pembrolizumab + carboplatin + paclitaxel were some of the drug combinations used as first-line agents in patients with lung cancer (Fig. 2b). Cyclophosphamide + doxorubicin and Docetaxel + pertuzumab were the drug combinations used in primary line of treatment for patients with breast cancer (Fig. 2c). Treatment lines generated by the algorithm were confirmed by two evaluators and were thus approved.
Pharmacological treatment for patients with lung cancer (n = 85) and breast cancer (n = 30) (a) Patients receiving pharmacological treatment for lung cancer as per the line of treatment (n = 85) (b) Common drug regimen in first-line (blue bars), second-line (red bars) and third-line (black bars) treatment for patients with lung cancer (n = 85). (c) Common drug regimen in first-line (blue bars), second-line (red bars) and third-line (black bars) treatment for patients with breast cancer (n = 30). The drug or combination of drugs for which the frequency is > 3 is presented here. aGenetic combination. CPA, cyclophosphamide; FU, fluorouracil; BVZ, bevacizumab; CBP, carboplatin; PTXL, paclitaxel; ATZ, atezolizumab.; PS, pemetrexed sodium
3.3 Adjudication results
Of 2,039 records in clinicians’ progress notes, 131 records in radiology reports and 60 records in pathological reports of 15 patients with lung cancer, treatment response was adjudicated by two evaluators. Among these records, 182 and 47 records, respectively, were adjudicated for OR, SD, or PD because other records were not related to treatment response (Online Resource, Supplementary Table 1). Among 28 therapy treatment lines used in 15 patients, best response to each treatment was identified. For the best response, clinicians’ progress notes were the most common primary source data for treatment response assessment (60.7%), followed by radiation reports (28.6%) (Fig. 3). A concordance correlation coefficient (kappa coefficient) of 0.59 was obtained for two evaluators in adjudicating the results of best response (Table 2).
3.4 Key terms extracted by evaluators for adjudication on response to treatment
Key terms were extracted by evaluators after adjudication of text data from 182 records in progress notes and 47 records in radiation reports. Among these, OR had 72 records, SD had 71 records and PD had 48 records in progress notes; and corresponding numbers in radiation reports were 26, 20, and 16. In total, 610 key terms were extracted from progress notes and 555 from radiation reports. Key terms were translated from Japanese to English. For treatment response of OR, “reduction/shrink” was the most common key term (69%) used to describe tumor response, with the highest sensitivity and specificity in the progress notes (Fig. 4a). However, it was less frequent in radiology reports (Online Resource, Supplementary Fig. 1a). “(No) remarkable change/ (no) aggravation)” was the most common key term (51–44%) used to describe stable disease in progress notes (Fig. 4b). Pertinent key terms used in radiology reports were “(no) lesion” and “(no) change” (Online Resource, Supplementary Fig. 1b). Key terms used to describe progressive disease were “(limited) effect” and “enlargement/grow” in progress notes (Fig. 4c). However, it was difficult to detect characteristic words in radiology reports (Online Resource, Supplementary Fig. 1). “reduction/shrink”, “effect” and “improvement” were mostly used in the positive context in reporting tumor response in progress notes and radiology reports. Whereas “remarkable change” was used in a negative context in progress reports and radiology reports (Table 3). Key terms identified in 15 patients were also confirmed in progress notes and radiology reports in the other larger cohort of patients with lung cancer not assessed by evaluators and also in patients with breast cancer (Online Resource, Supplementary Tables 2 and 3). No remarkable differences were found in the other larger cohort of patients with lung cancer. However, in the progress notes of the breast cancer patients, there was a little use of the terms related to lung such as “Infiltration shadow” and “Pleural effusion” that were identified as the key word in the lung patients. But again, these terms were used in the radiology reports of the patients with breast cancer.
Frequency, sensitivity and specificity of top 15 key terms for the adjudication extracted by the evaluators in progress notes (a) Treatment responses (n = 72 units: records) (b) Stable disease (n = 71 units: records) (c) Progression of disease (n = 48 units: records) (d) Sensitivity and specificity of key terms for treatment responses (e) Sensitivity and specificity of key terms for stable disease (f) Sensitivity and specificity of key terms for progression of disease
3.5 Mortality assessment
Throughout the study period, 3 deaths were recorded and all of them occurred in the hospital (2 deaths in patients with lung cancer and 1 in patient with breast cancer) (Online Resource, Supplementary Table 4).
4 Discussion
Our study demonstrated that evaluation of treatment response to each line of treatment was feasible in patients with lung cancer, using Japanese EHR such as progress notes and radiology reports that are in Japanese language. We identified key terms with high specificity and sensitivity to assess treatment response and we could also determine their use in positive/negative context.
In a study by Griffith et al., owing to missing data and lack of clarity in radiology reports, RECIST could not adequately assess progression of non-small cell lung cancer in EHR-derived data [8]. Our study aimed to assess response to drug treatment in clinical practice by taking into consideration the principle of the RECIST criteria including the decision of continuing/discontinuing treatment. It was feasible to evaluate the treatment response using criteria defined in this study which were based on retrospective review of radiology reports and progress notes. This approach of assessing the real-world response is increasingly used for comparative effectiveness research such as overall survival and progression-free survival particularly in the US but has not been done in Japan yet [9, 10, 14]. Our study provides the results of research evaluating such an approach using Japanese EHRs.
A limited number of studies has evaluated the feasibility of developing NLP tools for identifying textual sources for oncological outcomes in medical records regarding pharmacotherapy and tumor response [13, 14]. In these studies, NLP was employed to extract clinically relevant oncologic endpoints from unstructured EHR data [13, 14]. In this study we utilized NLP tools specialized for Japanese language for assessing response to drug treatment in patients with cancer. There were challenges pertaining to the use of non-English language in many other countries for using the NLP tools such as word segmentation and functional expression of Japanese language [15, 16]. This could be due to spelling variants and orthographical variants of Japanese language. For tackling these challenges in our study, the dictionary specialized for treatment responses in cancer patients was created on the top of the standard dictionary, which led high sensitivity and specificity of key terms related to treatment response. We also generated the rules of specifying positive/negative expressions for key terms broken down by morphological analysis, which was required for assessing response.
Our study also investigated the algorithm for lines of treatment of anticancer drugs. Patients with cancer in general receive several treatment lines and some of the regimens consist of multiple drugs with varying timing of drug administration. The information about treatment lines or treatment regimen is critical for ensuring drug effectiveness and safety in studies using large databases. Real-world treatment pattern is one of the frequently researched questions using administrative databases or EHR. Despite availability of prescription records/data, these databases have limited information regarding treatment line. However, some studies have reported methods to identify the treatment regimen using these databases and the algorithm for identifying them in our study is simple and feasible for large databases [22].
Our study has some inherent limitations. As the evaluators adjudicated fewer patients, since the purpose of this study is feasibility, we could not cross-validate the developed NLP rules. In addition, patients who had longer follow-up of anticancer drug therapy was selected for adjudication to evaluate diverse expressions for treatment responses of OR, SD, and PD, which might introduce selection bias, e.g., patients who have early treatment failure due to tumour flare or intolerance might not be presented among those 15 patients. Therefore, further studies are required to validate methodologies developed in our study. One of the important limitations of the Japanese EHR is the lack of linkage among hospitals and clinics in the Japanese medical practice environment resulting in the inability to analyze survival time/death outcomes. Moreover, treatment and imaging tests outside the hospital were not considered. Information about death and death date (confirmed date) is available in the city government database but could not be accessed for analysis purposes due to the personal information protection act. However, “Act on Anonymized Medical Data That Are Meant to Contribute to Research and Development in the Medical Field” (Next Generation Medical Infrastructure Law) may address the current issue of data accessibility and the aforementioned EHR “Millennial Medical Record Project” is the first organization certified by this low. Since the positive/negative context of key terms is important for adjudication of treatment response and for increasing the accuracy, we plan to investigate the utilization of AI/machine learning for the same. Transparency about the process of collecting the unstructured data for outcomes is also important [23]. Although the method involved double review by two independent reviews to minimize errors in identifying key terms, there was a possibility of AI identifying other key terms because of the large numbers of records to be reviewed. Source of EHR data was limited to a university hospital in this study. Future studies using EHR from several other hospitals across Japan would add diversity to the patient population.
This is the first real-world EHR database study that assesses response to anticancer treatment using Japanese EHRs; mainly progress notes and radiology reports. Findings from our study may form the base for future comparative efficacy research using real-world oncology outcomes in Japan. Within the limitations of our study, results showed that Japanese key terms for treatment response were identifiable by NLP. However, future studies using large scale EHR databases from several other hospitals in Japan are required to substantiate our results.
Data Availability
Data from the University of Miyazaki Hospital supporting the findings of this study may be made available upon request and are subject to approval by the ethics committee of University of Miyazaki.
References
Garrison LP Jr, Neumann PJ, Erickson P, Marshall D, Mullins CD. Value Health. 2007;10:326–35. https://doi.org/10.1111/j.1524-4733.2007.00186.x. Using real-world data for coverage and payment decisions: the ISPOR Real-World Data Task Force report.
Katkade VB, Sanders KN, Zou KH. Real world data: an opportunity to supplement existing evidence for the use of long-established medicines in health care decision making. J Multidiscip Healthc. 2018;11:295–304. https://doi.org/10.2147/jmdh.s160029.
Ehrenstein V, Kharrazi H, Lehmann H et al. (2019) Obtaining data from electronic health records, tools and technologies for registry interoperability, registries for evaluating patient outcomes: a user’s guide, 3rd Edition, Addendum 2 [Internet], Agency for Healthcare Research and Quality (US).
Kim E, Rubinstein SM, Nead KT, Wojcieszynski AP, Gabriel PE, Warner JL. The evolving use of electronic health records (EHR) for research. Semin Radiat Oncol. 2019;29:354–61. https://doi.org/10.1016/j.semradonc.2019.05.010.
Zarrinpar A, Cheng T-YD, Huo Z. What can we learn about drug safety and other effects in the era of electronic health records and big data that we would not be able to learn from classic epidemiology? J Surg Res. 2020;246:599–604. https://doi.org/10.1016/j.jss.2019.09.053.
Chapman WW, Nadkarni PM, Hirschman L, D’Avolio LW, Savova GK, Uzuner O. Overcoming barriers to NLP for clinical text: the role of shared tasks and the need for additional creative solutions. J Am Med Inform Assoc. 2011;18:540–3. https://doi.org/10.1136/amiajnl-2011-000465.
Capurro D, Yetisgen M, van Eaton E, Black R, Tarczy-Hornoch P. Availability of structured and unstructured clinical data for comparative effectiveness research and quality improvement: a multisite assessment. EGEMS (Wash DC). 2014;2:1079. https://doi.org/10.13063/2327-9214.1079.
Griffith SD, Tucker M, Bowser B, et al. Generating real-world tumor burden endpoints from electronic health record data: comparison of RECIST, radiology-anchored, and clinician-anchored approaches for abstracting real-world progression in non-small cell lung cancer. Adv Ther. 2019;36:2122–36. https://doi.org/10.1007/s12325-019-00970-1.
Khozin S, Miksad RA, Adami J, et al. Real-world progression, treatment, and survival outcomes during rapid adoption of immunotherapy for advanced non-small cell lung cancer. Cancer. 2019;125:4019–32. https://doi.org/10.1002/cncr.32383.
Ma X, Bellomo L, Magee K, et al. Characterization of a real-world response variable and comparison with RECIST-based response rates from clinical trials in advanced NSCLC. Adv Ther. 2021;38:1843–59. https://doi.org/10.1007/s12325-021-01659-0.
DeMichele A, Cristofanilli M, Brufsky A, et al. Comparative effectiveness of first-line palbociclib plus letrozole versus letrozole alone for HR+/HER2 – metastatic breast cancer in US real-world clinical practice. Breast Cancer Res. 2021;23:37. https://doi.org/10.1186/s13058-021-01409-8.
Bagley SJ, Talento S, Mitra N, Meropol NJ, Cohen RB, Langer CJ, Vachani A. Comparative effectiveness of carboplatin/pemetrexed with versus without bevacizumab for advanced nonsquamous non–small cell lung cancer. J Natl Compr Canc Netw. 2019;17:469–77. https://doi.org/10.6004/jnccn.2018.7102.
Li Y, Luo Y-H, Wampfler JA, et al. Efficient and accurate extracting of unstructured EHRs on cancer therapy responses for the development of RECIST natural language processing tools: part I, the corpus. JCO Clin Cancer Inform. 2020;4:383–91. https://doi.org/10.1200/cci.19.00147.
Kehl KL, Elmarakeby H, Nishino M, et al. Assessment of deep natural language processing in ascertaining oncologic outcomes from radiology reports. JAMA Oncol. 2019;5:1421–9. https://doi.org/10.1001/jamaoncol.2019.1800.
Névéol A, Dalianis H, Velupillai S, Savova G, Zweigenbaum P. Clinical natural language processing in languages other than english: opportunities and challenges. J Biomed Semantics. 2018;9:12. https://doi.org/10.1186/s13326-018-0179-8.
Matsuyoshi S, Sato S, Utsuro T. Compilation of a dictionary of japanese functional expressions with hierarchical organization. In: Matsumoto Y, Sproat RW, Wong KF, Zhang M, editors. Computer processing of oriental languages. Beyond the orient: the research challenges ahead. Berlin, Heidelberg: Springer; 2006. pp. 395–402.
Initiative LD, Millennial Medical Record Project. https://www.ldi.or.jp/. Accessed: 26 September 2021.
Eisenhauer EA, Therasse P, Bogaerts J, et al. New response evaluation criteria in solid tumours: revised RECIST guideline (version 1.1). Eur J Cancer. 2009;45:228–47. https://doi.org/10.1016/j.ejca.2008.10.026.
NTT DATA Mathematical Systems Inc. HP. https://www.msi.co.jp/tmstudio/. Accessed: 03 December 2021.
Medical Information System Development Center (MEDIS-DC). http://www.medis.or.jp/. Accessed: 1 July 2022.
New Guidelines to Evaluate the Response to Treatment in Solid Tumors (RECIST Guidelines). - Revised Version 1.1 - JCOG Version Version 1.0 http://www.jcog.jp/doctor/tool/recistv11.html. Accessed: 1 July 2022.
Palmaro A, Gauthier M, Despas F, Lapeyre-Mestre M. Identifying cancer drug regimens in french health insurance database: an application in multiple myeloma patients. Pharmacoepidemiol Drug Saf. 2017;26:1492–9. https://doi.org/10.1002/pds.4266.
Wang SV, Patterson OV, Gagne JJ, et al. Transparent reporting on research using unstructured electronic health record data to generate ‘real world’evidence of comparative effectiveness and safety. Drug Saf. 2019;42:1297–309. https://doi.org/10.1007/s40264-019-00851-0.
Acknowledgements
Editorial and/or medical writing support was provided by MedPro Clinical Research (Haruyoshi Ogata), supported by CBCC Global Research (Leena Patel), and was funded by Pfizer Japan Inc.
Funding
The study was sponsored by Pfizer Japan Inc.
Author information
Authors and Affiliations
Contributions
All authors contributed to the concept and design of the study, analysis and interpretation of data, drafting/revising the manuscript for important intellectual content, and approval of the final version to be published.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
This study was approved by the ethics committee of University of Miyazaki (registration no.: 0-0845) and the opt-out consent process was granted. The study was conducted in accordance with the Ethical Guidelines for Medical and Health Research Involving Human Subjects by Ministry of Education, Culture, Sports, Science and Technology (MEXT), and Ministry of Health, Labor and Welfare (MHLW).
Consent for publication
All authors agreed with the content and that all gave explicit consent to submit the final version to be published.
Competing Interests
Kenji Araki and Nobuhiro Matsumoto have received research funding from Pfizer.
Kanae Togo, Naohiro Yonemoto, Emiko Ohki and Linghua Xu are employees of Pfizer Japan Inc. Taiga Miyazaki is consultant or advisor in Pfizer, Kyorin, and Asahi Kasei Pharma.
Taiga Miyazaki is also a Speakers’ bureau in Pfizer, MSD, Astellas, Sumitomo Dainippon Pharma, Kyorin, GSK, Lilly, Meiji Seika Pharma, Novartis, Boehringer Ingelheim, and AstraZeneca. Taiga Miyazaki has also received research funding from Pfizer, Astellas, Toyama Chemical, Asahi Kasei Pharma, and MSD.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Araki, K., Matsumoto, N., Togo, K. et al. Real-world treatment response in Japanese patients with cancer using unstructured data from electronic health records. Health Technol. 13, 253–262 (2023). https://doi.org/10.1007/s12553-023-00739-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12553-023-00739-1