Abstract
Purpose
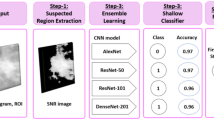
Breast cancer (BC) is the most common diagnosed cancer type and one of the top leading causes of death in women worldwide. This paper aims to investigate ensemble learning and transfer learning for binary classification of BC histological images over the four-magnification factor (MF) values of the BreakHis dataset: 40X, 100X, 200X, and 400X.
Methods
The proposed homogeneous ensembles are implemented using a hybrid architecture that combines: (1) three of the most recent deep learning (DL) techniques for feature extraction: DenseNet_201, MobileNet_V2, and Inception_V3, and (2) four of the most popular boosting methods for classification: AdaBoost (ADB), Gradient Boosting Machine (GBM), LightGBM (LGBM) and XGBoost (XGB) with Decision Tree (DT) as a base learner. The study evaluated and compared: (1) a set of boosting ensembles designed with the same hybrid architecture and different number of trees (50, 100, 150 and 200); (2) different boosting methods, and (3) the single DT classifier with the best boosting ensembles. The empirical evaluations used: four classification performance criteria (accuracy, recall, precision and F1-score), the fivefold cross-validation, Scott Knott statistical test to select the best cluster of the outperforming models, and Borda Count voting system to rank the best performing ones.
Results
The best boosting ensemble achieved an accuracy value of 92.52% and it was constructed using XGB with 200 trees and Inception_V3 as feature extractor (FE).
Conclusions
The results showed the potential of combining DL techniques for feature extraction and boosting ensembles to classify BC in malignant and benign tumors.








Similar content being viewed by others
References
Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, Bray F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin. 2021;71:209–49. https://doi.org/10.3322/caac.21660.
Ginsburg O, Yip C-H, Brooks A, Cabanes A, Caleffi M, Yataco JAD, Gyawali B, McCormack V, de Anderson MM, Mehrotra R, Mohar A, Murillo R, Pace LE, Paskett ED, Romanoff A, Rositch AF, Scheel JR, Schneidman M, Unger-Saldaña K, Vanderpuye V, Wu T-Y, Yuma S, Dvaladze A, Duggan C, Anderson BO. Breast cancer early detection: a phased approach to implementation. Cancer. 2020;126:2379–93. https://doi.org/10.1002/cncr.32887.
Hela B, Hela M, Kamel H, Sana B, Najla M. Breast cancer detection: a review on mammograms analysis techniques. In: 10th International Multi-Conferences on Systems, Signals Devices 2013 (SSD13); 2013. p. 1–6. https://doi.org/10.1109/SSD.2013.6563999.
Bukhari MH, Akhtar ZM. Comparison of accuracy of diagnostic modalities for evaluation of breast cancer with review of literature. Diagn Cytopathol. 2009;37:416–24. https://doi.org/10.1002/dc.21000.
Giess CS, Frost EP, Birdwell RL. Difficulties and errors in diagnosis of breast neoplasms. Semin Ultrasound CT MRI. 2012;33:288–99. https://doi.org/10.1053/j.sult.2012.01.007.
Abdar M, Zomorodi-Moghadam M, Zhou X, Gururajan R, Tao X, Barua PD, Gururajan R. A new nested ensemble technique for automated diagnosis of breast cancer. Pattern Recogn Lett. 2020;132:123–31. https://doi.org/10.1016/j.patrec.2018.11.004.
Yassin NIR, Omran S, El Houby EMF, Allam H. Machine learning techniques for breast cancer computer aided diagnosis using different image modalities: a systematic review. Comput Methods Programs Biomed. 2018;156:25–45. https://doi.org/10.1016/j.cmpb.2017.12.012.
Hamed G, Marey MAE-R, Amin SE-S, Tolba MF. Deep learning in breast cancer detection and classification. In: Hassanien A-E, Azar AT, Gaber T, Oliva D, Tolba FM, editors. Proceedings of the International Conference on Artificial Intelligence and Computer Vision (AICV2020). Springer International Publishing, Cham; 2020. p. 322–33. https://doi.org/10.1007/978-3-030-44289-7_30.
Schelter S, Biessmann F, Januschowski T, Salinas D, Seufert S, Szarvas G. On challenges in machine learning model management. 11.
Bateman B, Jha AR, Johnston B, Mathur I. The supervised learning workshop: a new, interactive approach to understanding supervised learning algorithms. 2nd ed. Packt Publishing; 2020.
Breiman L. Bagging predictors. Mach Learn. 1996;24:123–40. https://doi.org/10.1007/BF00058655.
Kong EB, Dietterich TG. Error-correcting output coding corrects bias and variance. In: Machine Learning Proceedings. Elsevier; 1995. p. 313–21. https://doi.org/10.1016/B978-1-55860-377-6.50046-3.
Ganaie MA, Hu M, Tanveer M, Suganthan PN. Ensemble deep learning: a review. arXiv:2104.02395 [cs]. 2021.
El Ouassif B, Idri A, Hosni M. Homogeneous ensemble based support vector machine in breast cancer diagnosis: In: Proceedings of the 14th International Joint Conference on Biomedical Engineering Systems and Technologies. SCITEPRESS - Science and Technology Publications, Online Streaming, --- Select a Country; 2021. p. 352–60. https://doi.org/10.5220/0010230403520360.
Mayr A, Binder H, Gefeller O, Schmid M. The evolution of boosting algorithms - from machine learning to statistical modelling. Methods Inf Med. 2014;53:419–27. https://doi.org/10.3414/ME13-01-0122.
Zhou Z-H. Ensemble learning. In: Li SZ, Jain A, editors. Encyclopedia of Biometrics. Springer US, Boston, MA; 2009. p. 270–3. https://doi.org/10.1007/978-0-387-73003-5_293.
Hosni M, Abnane I, Idri A, Carrillo de Gea JM, Fernández Alemán JL. Reviewing ensemble classification methods in breast cancer. Comput Methods Programs Biomed. 2019;177;89–112. https://doi.org/10.1016/j.cmpb.2019.05.019.
Bühlmann P, Yu B. Analyzing bagging. Ann Statist. 2002;30. https://doi.org/10.1214/aos/1031689014.
Koltchinskii V, Yu B. Three papers on boosting: an introduction. Ann Stat. 2004;32:12–12. https://doi.org/10.1214/aos/1079120127.
Opitz D, Maclin R. Popular ensemble methods: an empirical study. JAIR. 1999;11:169–98. https://doi.org/10.1613/jair.614.
Tuv E. Ensemble learning. In: Guyon I, Nikravesh M, Gunn S, Zadeh LA (eds.). Feature extraction: Foundations and applications. Springer, Berlin, Heidelberg; 2006. p. 187–204. https://doi.org/10.1007/978-3-540-35488-8_8.
Rahman S, Irfan M, Raza M, Moyeezullah Ghori K, Yaqoob S, Awais M. Performance Analysis of Boosting Classifiers in Recognizing Activities of Daily Living. Int J Environ Res Public Health. 2020;17:1082. https://doi.org/10.3390/ijerph17031082.
Sutton CD. Classification and regression trees, bagging, and boosting. In: Handbook of Statistics. Elsevier; 2005. p. 303–29. https://doi.org/10.1016/S0169-7161(04)24011-1.
Theodoridis G, Tsadiras A. Using machine learning methods to predict subscriber churn of a web-based drug information platform. In: Maglogiannis I, Macintyre J, Iliadis L, editors. Artificial Intelligence Applications and Innovations. Springer International Publishing, Cham; 2021. p. 581–93. https://doi.org/10.1007/978-3-030-79150-6_46.
Zerouaoui H, Idri A. Deep hybrid architectures for binary classification of medical breast cancer images. Biomed Signal Process Control. 2022;71: 103226. https://doi.org/10.1016/j.bspc.2021.103226.
del Rio F, Messina P, Dominguez V, Parra D. Do better imagenet models transfer better... for image recommendation? arXiv:1807.09870 [cs]. (2018).
Wang S-H, Zhang Y-D. DenseNet-201-Based deep neural network with composite learning factor and precomputation for multiple sclerosis classification. ACM Trans Multimedia Comput Commun Appl. 2020;16:1–19. https://doi.org/10.1145/3341095.
Howard A, Zhmoginov A, Chen L-C, Sandler M, Zhu M. Inverted residuals and linear bottlenecks: mobile networks for classification, detection and segmentation. In: CVPR. 2018.
Iqbal M, Yan Z. Supervised machine learning approaches: a survey. Int J Soft Comput. 2015;5:946–52. https://doi.org/10.21917/ijsc.2015.0133.
Hastie T, Tibshirani R, Friedman J. Ensemble learning. In: Hastie T, Tibshirani R, Friedman J, editors. The elements of statistical learning: Data mining, inference, and prediction. Springer, New York, NY; 2009. p. 605–24. https://doi.org/10.1007/978-0-387-84858-7_16.
Cao D-S, Xu Q-S, Liang Y-Z, Zhang L-X, Li H-D. The boosting: a new idea of building models. Chemom Intell Lab Syst. 2010;100:1–11. https://doi.org/10.1016/j.chemolab.2009.09.002.
Cao Y, Miao Q-G, Liu J-C, Gao L. Advance and prospects of AdaBoost algorithm. Acta Automatica Sinica. 2013;39:745–58. https://doi.org/10.1016/S1874-1029(13)60052-X.
Schapire RE. Explaining AdaBoost. In: Schölkopf B, Luo Z, Vovk V, editors. Empirical inference: Festschrift in honor of Vladimir N. Vapnik. Springer, Berlin, Heidelberg; 2013. p. 37–52. https://doi.org/10.1007/978-3-642-41136-6_5.
Ayyadevara VK. Gradient boosting machine. In: Ayyadevara VK, editor. Pro machine learning algorithms : a hands-on approach to implementing algorithms in Python and R. Apress, Berkeley, CA; 2018. p. 117–34. https://doi.org/10.1007/978-1-4842-3564-5_6.
Chen T, He T. xgboost: eXtreme gradient boosting. 4.
Chen T, Guestrin C. XGBoost: a scalable tree boosting system. In: Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining. ACM, San Francisco California USA; 2016. p. 785–94. https://doi.org/10.1145/2939672.2939785.
Ke G, Meng Q, Finley T, Wang T, Chen W, Ma W, Ye Q, Liu T-Y. LightGBM: a highly efficient gradient boosting decision tree. 9.
Welcome to LightGBM’s documentation!. LightGBM 3.2.1.99 documentation. https://lightgbm.readthedocs.io/en/latest/index.html. Accessed 6 Jun 2021.
Sadoughi F, Kazemy Z, Hamedan F, Owji L, Rahmanikatigari M, Azadboni TT. Artificial intelligence methods for the diagnosis of breast cancer by image processing: a review. Breast Cancer (Dove Med Press). 2018;10:219–30. https://doi.org/10.2147/BCTT.S175311.
Zerouaoui H, Idri A. Reviewing machine learning and image processing based decision-making systems for breast cancer imaging. J Med Syst. 2021;45:8. https://doi.org/10.1007/s10916-020-01689-1.
Logan R, Williams BG, Ferreira da Silva M, Indani A, Schcolnicov N, Ganguly A, Miller SJ. Deep convolutional neural networks with ensemble learning and generative adversarial networks for alzheimer’s disease image data classification. Front Aging Neurosci. 2021;13:497. https://doi.org/10.3389/fnagi.2021.720226.
Kassani SH, Kassani PH, Wesolowski MJ, Schneider KA, Deters R: Classification of histopathological biopsy images using ensemble of deep learning networks. arXiv:1909.11870 [cs, eess]. 2019.
Vo DM, Nguyen N-Q, Lee S-W. Classification of breast cancer histology images using incremental boosting convolution networks. Inf Sci. 2019;482:123–38. https://doi.org/10.1016/j.ins.2018.12.089.
Osman A, Aljahdali HM. An effective of ensemble boosting learning method for breast cancer virtual screening using neural network model. IEEE Access. 2020. https://doi.org/10.1109/ACCESS.2020.2976149.
Zerouaoui H, Idri A, Nakach FZ, Hadri RE. Breast fine needle cytological classification using deep hybrid architectures. In: Gervasi O, Murgante B, Misra S, Garau C, Blečić I, Taniar D, Apduhan BO, Rocha AMAC, Tarantino E, Torre CM, editors. Computational science and its applications – ICCSA 2021. Springer International Publishing, Cham; 2021. p. 186–202. https://doi.org/10.1007/978-3-030-86960-1_14.
Spanhol FA, Oliveira LS, Petitjean C, Heutte L. A dataset for breast cancer histopathological image classification. IEEE Trans Biomed Eng. 2016;63:1455–62. https://doi.org/10.1109/TBME.2015.2496264.
Breast Cancer Histopathological Database (BreakHis). Laboratório Visão Robótica e Imagem. https://web.inf.ufpr.br/vri/databases/breast-cancer-histopathological-database-breakhis/. Accessed 12 May 2021.
B, N. Image data pre-processing for neural networks. https://becominghuman.ai/image-data-pre-processing-for-neural-networks-498289068258. Accessed 12 May 2021.
Yussof W. Performing contrast limited adaptive histogram equalization technique on combined color models for underwater image enhancement. 2013.
Shorten C, Khoshgoftaar TM. A survey on image data augmentation for deep learning. J Big Data. 2019;6:60. https://doi.org/10.1186/s40537-019-0197-0.
Jelihovschi E, Faria JC, Allaman IB. ScottKnott: a package for performing the Scott-Knott clustering algorithm in R. Tend Mat Apl Comput. 2014;15:003. https://doi.org/10.5540/tema.2014.015.01.0003.
Emerson P. The original Borda count and partial voting. Soc Choice Welf. 2013;40:353–8. https://doi.org/10.1007/s00355-011-0603-9.
Guo Y, Shi H, Kumar A, Grauman K, Rosing T, Feris R. SpotTune: Transfer learning through adaptive fine-tuning. 10.
Pan SJ, Yang Q. A survey on transfer learning. IEEE Trans Knowl Data Eng. 2010;22:1345–59. https://doi.org/10.1109/TKDE.2009.191.
The Elements of Statistical Learning. SpringerLink. https://link.springer.com/book/10.1007/978-0-387-84858-7. Accessed 1 Jun 2021.
Kuhn M, Johnson K. Applied predictive modeling. Springer New York, New York, NY; 2013. https://doi.org/10.1007/978-1-4614-6849-3.
Funding
This work was conducted under the research project “Machine Learning based Breast Cancer Diagnosis and Treatment”, 2020–2023. The authors would like to thank the Moroccan Ministry of Higher Education and Scientific Research, Digital Development Agency (ADD), CNRST, and UM6P for their support.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Research involving human participants and/or animals
NA.
Informed consent
NA.
Conflicts of interest
The authors declare that they have no conflicts of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Nakach, FZ., Zerouaoui, H. & Idri, A. Hybrid deep boosting ensembles for histopathological breast cancer classification. Health Technol. 12, 1043–1060 (2022). https://doi.org/10.1007/s12553-022-00709-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12553-022-00709-z