Abstract
Raman spectroscopy offers the potential for fingerprinting biological molecules at ultra-low concentration and therefore has potential for the detection of viruses. Here we review various Raman techniques employed for the investigation of viruses. Different Raman techniques are discussed including conventional Raman spectroscopy, surface-enhanced Raman spectroscopy, Raman tweezer, tip-enhanced Raman Spectroscopy, and coherent anti-Stokes Raman scattering. Surface-enhanced Raman scattering can play an essential role in viral detection by multiplexing nanotechnology, microfluidics, and machine learning for ensuring spectral reproducibility and efficient workflow in sample processing and detection. The application of these techniques to diagnose the SARS-CoV-2 virus is also reviewed.
Graphical abstract

Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Viruses, described as “organisms at the edge of life,” are known to be the causative agents of most of the human pandemics reported in history (Rybicki 1990). Viruses are infectious agents with submicroscopic sizes which are obligate intercellular parasites. The traditional methods of viral detection rely on viral isolation and culture techniques such as plaque assay, hemagglutination assay, and histological observations (Hematian et al. 2016; Storch 2000). However, these techniques demand skilled labor, a culture-based approach, and longer duration. Direct examination techniques such as electron microscopy or immunofluorescence usually suffer from poor sensitivity and specificity and the data can be difficult to interpret (Burrell et al. 2017). Polymerase chain reaction (PCR) techniques have been employed since the 1990s and represent the current gold standard (Espy et al. 2006). However, the high workload and reagent shortage during the epidemic makes testing during viral pandemics using the PCR technique a challenging task.
Spectroscopic modalities, Raman spectroscopy in particular, allow for detailed investigation of various kinds of biological samples, with one prominent example being cancer detection (Ibrahim et al. 2021). The use of Raman spectroscopy for viral detection from body fluids has seen a resurgence (Saleem et al. 2013; Saade et al. 2008; Naseer et al. 2019; Desai et al. 2020). Virus detection can be achieved either directly, by identifying the components of the virus, or indirectly by detecting components from the virus elicited immune response (Ramoji et al. 2022). The standard target biomolecules in the former case are glycoproteins, nucleocapsid, and viral genome; the typical target biomolecules for the latter case are antibodies and cytokines (Yang et al. 2021). The change in relative distribution of various components such as proteins, amino acids, lipids, and carbohydrate derivatives in body fluids can also act as an indicator of viral infection. Capturing such biochemical signatures in the nanoscale regime can be realized by versatile Raman spectroscopic techniques such as surface-enhanced Raman spectroscopy (SERS), Raman tweezers, tip-enhanced Raman spectroscopy (TERS), and coherent anti-Stokes Raman scattering (CARS). Recently, Savinon-Flores et al. have reviewed SERS-based methods for the detection of viral infections in humans (Savinon-Flores et al. 2021). The present study extends this discussion to other variants of Raman spectroscopy.
Raman spectroscopy
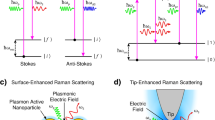
Raman spectroscopy is a technique used for the determination of molecular structure based on vibrational frequencies of bonds by analyzing the light scattered from the sample of interest (Das and Agrawal 2011). This technique utilizes the change in polarizability of the molecular species upon irradiation with electromagnetic radiation and a concomitant inelastic scattering to fingerprint the chemical identity of the molecule. The inelastically scattered light (Raman scattering) produced by excitation with monochromatic radiation contains rich information about molecular vibrations which has led to it becoming a popular technique to analyze many kinds of biological samples (e.g., tissues and body fluids) (Dietzek et al. 2010). The efficiency of Raman scattering is typically very low, such that it accounts for only 1 in 10 million incident photons. In Stokes Raman scattering, the scattered radiation has lower energy compared to the incident radiation, whereas, in anti-Stokes Raman scattering, the scattered radiation has higher energy than that of the incident light (Popp and Kiefer 2006). The schematic of the Raman scattering process is shown in Fig. 1 (George 2020). The Stokes scattering is more favored as compared to the anti-Stokes scattering under ambient conditions. In Raman measurements, the dominant Rayleigh scattered radiation is filtered out to obtain the Raman signal from the sample of interest. The popularity of Raman spectroscopy for investigating biological samples can be attributed to the insignificant Raman scattering of the water, which is the major component in almost all biological entities. This technique does not require expensive solvents, matrix, sample processing steps, or lengthy data analysis time.
Conceptual representation of scattering process (George 2020)
Advancements in optical instrumentation have facilitated the usage of the Raman technique in a wide range of biomedical applications. These developments have involved improvements in light sources and detectors relating to diode lasers, CCDs, mini-spectrometers, and optical fiber probes (Zhu et al. 2014). Nanotechnology-based developments in Raman spectroscopy has led to SERS, which has allowed for ultra-sensitive detection of biomolecular analytes when present at very low concentrations in body fluids (Culha et al. 2012).
Raman spectroscopy in viral diagnostics
Biosensors exploiting the Raman effect have been reported for a variety of viruses, and a few are discussed here. Micro-Raman spectroscopy has been used to investigate adenovirus-infected human embryonic kidney epithelial (HEK293) cells (Moor et al. 2014). The study was capable of detecting viruses within cells from the spectra recorded after 12 h of viral infection, which is comparatively quicker than the routinely adopted immunostaining technique. The researchers identified one Raman spectral line at around 952 cm−1 that can be used as a spectral marker for identifying virus proliferation in cells. In 2018, the same group demonstrated the detection of adenovirus in the human cell within 3 h of viral infection (Moor et al. 2018). They suggested that two mechanisms occur in the cell following virus invasion. An initial reaction, proceeding without any protein production or nucleic acid replication, occurs immediately after viral entry, and a later phase in which both nucleic acid replication and viral protein production occur. Raman spectra were shown to be effective in identifying the compositional changes in DNA and RNA due to the cellular response upon viral invasion. In another work, the Raman technique was used to detect herpes virus in cell cultures (Salman et al. 2014). These experiments conducted on Vero cells were able to discriminate between control and herpes simplex virus type 1 (HSV-1) infected cases with 100% sensitivity. The major spectral contributions were exhibited in the amide III region comprising bands arising from proteins, lipids, and nucleic acids. Raman spectroscopy has been also employed for evaluating the biochemical changes induced due to Epstein–Barr virus infection in human glial cells (Tiwari et al. 2020). That study found differences in the Raman spectra obtained from the nucleus and periphery, which indicates that the viral invasion and its progression have resulted in different signaling processes at these locations. Combined with multivariate analysis, Raman techniques have been used to detect porcine parvovirus from cell cultures with a sensitivity and accuracy of 97.77% and 99.97%, respectively (Gogone et al. 2021).
Saleem et al. have applied the Raman technique to whole blood and serum samples for the detection of dengue viral infection. The experiments conducted using both 532 nm and 442 nm laser excitations were able to identify the dengue infection from the Raman marker bands at 1614 cm−1 and 1750 cm−1. These bands were originated from the presence of antibodies (IgG and IgM) in blood samples induced in response to viral infection (Rehman et al. 2012). Later, the same group employed partial least square (PLS) regression modeling on the Raman measurements of infected serum samples achieving an accuracy of 100% when tested on six blind samples (Saleem et al. 2013). Bands (at 750 and 850 cm−1) corresponding to adenosine diphosphate (ADP) that arose from thrombocyte cell rupture were observed in dengue virus-infected cases. Besides, the presence of cortisone signature at 1732 cm−1 and the absence of carotenoid bands (1006, 1156, and 1516 cm−1) were found in the case of infected cells. Saade et al. have used the Raman technique to probe hepatitis virus in blood samples (Saade et al. 2008) by observing variation in the phospholipid band intensities which they attributed to enzymes triggered by hepatocytes damage in hepatitis. Naseer and his team differentiated between dengue and typhoid infections in human serum by use of the Raman spectroscopy technique (Naseer et al. 2019). This study was successful in identifying specific Raman biomarkers for both bacterial and viral infections from the serum sample. A band at 1686 cm−1 was proposed as a marker for dengue viral infection, which originated from soluble ST2 protein in dengue patients. In a recent pilot study, Desai et al. used Raman spectral features for the identification of viral RNA from saliva, with a sensitivity of 7.05 × 107 TU/mL (transduction units per mL) viral particles (Desai et al. 2020).
In a recent report, the automated deep learning-based TL-ResNet101 approach was employed to the analysis of Raman spectral images for identifying HBV infection from plasma (Ali et al. 2021). Deep transfer learning (DTL) was used to identify variations in infected Raman spectra and demonstrated high sensitivity (100%) as well as accuracy (99.7%) for hepatitis B virus (HBV) diagnosis. Another recent work used multiscale convolution neural networks (MCNN) for the classification of hepatitis infections from the Raman spectra of the serum sample (Zhao et al. 2021). Monsees et al. identified the Raman peak intensity ratio between nucleic acids and proteins useful for classification of virally infected cells from that of uninfected. They used Raman micro-spectroscopy for collecting the Raman signal from the phi6 (bacteriophage) infected Pseudomonas syringae and phi29 infected Bacillus subtilis as well as corresponding controls (Monsees et al. 2022). They observed an increase in the ratio for the former case and a decrease in the latter. Raman tweezers is a variant of Raman spectroscopy which is used in combination with optical tweezers to facilitate the investigation of live cells in physiological media without the requirement of any chemical/physical fixation. Raman tweezers use an optical trap (created by a tightly focused laser beam) to optically arrest the biological cell with simultaneous measurement of Raman spectra from the sample. This technique was used to study the influence of exogenous stress agents on human red blood cells. This includes monitoring the impact of extracellular tonicity, metallic nanoparticles, bisphenol A, alcohol, and normal saline on human red blood cells. We have previously demonstrated the use of Raman tweezers to investigate the response of single cells to external factors (Bankapur et al. 2010, 2014; Barkur et al. 2015; Lukose et al. 2019). Raman tweezers have also been used for the identification of Kaposi’s sarcoma-associated herpesvirus (KSHV) infected cells (BCBL-1 and BC-1 cells) from uninfected BJAB cells (Hamden et al. 2005). Raman tweezers were used to identify herpes viral infection based on Raman frequencies of proteins and nucleic acids at 1004, 1093, and 1664 cm−1 (Hamden et al. 2005). Pilat and team employed the Raman tweezer technique for analyzing the Staphylococcus aureus infected by two virulent phages JK2 and 80α, identifying band intensity at 746 cm−1 and at 1128 cm−1 attributed to heme required for energy production at the initial stage of infection (Pilat et al. 2020). Phage replication inside the host was shown by following Raman spectra intensity corresponding to the phosphodiester bond (1095 cm−1). As the host cells leak their cytoplasmic content during lysis, this leads to a decrease of cytosine and uracil peak at 782 cm−1 (Pilat et al. 2020). The corresponding Raman analysis scheme is shown in Fig. 2.
Bacteriophage invasion and corresponding molecular changes probed through Raman tweezer. Reprinted with permission from Pilat et al. (2020)
Cialla and colleagues demonstrated the applicability of TERS for viral investigations by extracting Raman signatures from the coat proteins and RNA of mosaic virus (Cialla et al. 2009). Ideally, the TERS technique combines the advantage of chemical sensitivity of the SERS technique with the high spatial resolution at the nanometer scale of a scanning probe technique. Nanoscale chemical imaging is realized in this technique with the aid of the electromagnetic field confined to the tip of a scanning probe microscope (SPM) (due to localized surface plasmon resonance) the so-called lightning-rod-effect (Cialla et al. 2009). The Abbe diffraction limit of the conventional Raman technique was shown to be circumvented by the single point TERS technique when used for investigating avipoxvirus (APV) and adeno-associated virus (AAV). Changes in the relative peak intensities as well as the position of spectral features were recorded for different particles from different virus strains (Hermann et al. 2011). In other recent work, a two-component approach employing TERS and AFM-IR techniques was used for both the identification as well as the structural characterization of individual virions (Dou et al. 2020). This combined approach was effective in determining protein secondary structure and amino acid composition of viral surfaces and was also able to probe the structural organization of individual virions of herpes simplex virus type 1 (HSV-1) and MS2 (Dou et al. 2020). Popp et al. used this technique together with chemometric analysis for the discrimination of varicella-zoster virus (VZV) and porcine teschovirus (PTV) with an accuracy of 91% (Olschewski et al. 2015). Single virus detection ability of this technique was demonstrated for classification of influenza virus and picornavirus (Deckert et al. 2020). The possibility of CARS technique for viral detection has been proposed, where the high electromagnetic field enhancement arising from the four-wave mixing of non-linear optics can overcome the limitations of weak inherent signals in the Raman technique (Deckert et al. 2020). The possibility of combining femtosecond adaptive spectroscopic techniques with enhanced resolution (FASTER)-CARS has been applied for individual viral particle detection (Deckert et al. 2020). Deckert et al. have achieved both quantitative and qualitative analyses at single virus level by effectively combining the tip-enhanced microscopy with coherent anti-Stokes Raman scattering (Fig. 3A). They were able to identify single copies of the H1N1 virus and coxsackie B3 virus solely based on the surface composition, as shown in Fig. 3B, C (Deckert et al. 2020). Detailed account of a single virus could be acquired since the tip is scanning the virus surface several times unlike the conventional SERS, where the averaging is done by investigating a large number of individual samples.
Schematics of identification of individual viruses (H1N1 and coxsackie virus) using TERS multiplexed with CARS. Reprinted from Deckert et al. (2020)
Resonance Raman spectroscopy (RRS), another variant of Raman spectroscopy, is a technique, where the incident laser energy is selected to be as close to the electronic transition energy of the sample (Butler et al. 2016). RRS has been effective in providing an enhancement factor of 102–106 for the Raman active vibrational frequency modes, which affords improvement of the detection limit (Butler et al. 2016; Wood and McNaughton 2006; Wood et al. 2004; Kuzmin et al. 2016). However, background fluorescence originating from the samples is a matter of concern in RRS. Use of UV-resonance Raman spectroscopy has been shown to be useful for analyzing virological samples, especially samples like filamentous viruses (Wen and Thomas 1998, Wen et al. 1999, 1997; Tadesse et al. 2020). Wen et al. have studied virus constituents using four different UV excitation wavelengths (257, 244, 238, and 229 nm) and were able to generate a databank for aromatic amino acids from proteins as well as for DNA nucleosides (Wen and Thomas 1998). Although identical signatures for coat protein tyrosine were found in filamentous viruses Pf1 and fd virions, the use of UV-resonance Raman spectroscopy indicated that the DNA organization was different (Wen et al. 1999). Supplementary Table 2 summarizes studies on detection of viruses of different kinds using Raman spectroscopy techniques.
Surface-enhanced Raman spectroscopy for virus detection
The use of conventional Raman spectroscopy for sensitive viral detection has been limited by weak signal strength (due to the low Raman scattering cross-section of biological samples). The low Raman scattering cross-section necessitates a longer signal acquisition time that often leads to sample damage (George 2020). This difficulty can be mitigated via the use of plasmonic nanostructures and consequent enhancement in the Raman signal, now popularly known as the surface-enhanced Raman spectroscopy (SERS) technique (Tadesse et al. 2020). Although resonance Raman spectroscopy can be applied for obtaining the fingerprint information of the molecule at very low concentration, the requirement for wavelength matching of excitation wavelength with electronic energy level of the molecule limits the technique. Moreover, most biomolecules exhibit absorption in the UV range and exposure to the radiation at this wavelength for long durations can damage clinical samples (Shipp et al. 2017). Despite the ongoing research on various biological systems using coherent anti-Stokes Raman scattering (CARS) technique, the increased complexity of the instrumentation precludes this technique as routine tool in clinical analysis. The application of tip-enhanced Raman spectroscopy (TERS) technique has been also restricted in biological sciences due to the complexity in fabricating reproducible tips (Huang et al. 2015). Raman tweezers have been associated with expensive and bulky instrumentation, which is unfavorable for point-of-care applications in clinical settings. Among the reported literature for SARS-CoV-2 detection using Raman-based techniques, the SERS technique has shown high promise for routine viral detection with satisfactory detection limit (George et al. 2018). The low Raman scattering cross-section, especially in the case of biological samples, can limit the applications of Raman spectroscopy for bioanalytical sensing purposes. To mitigate this, in SERS, the scattered light signal is improved by keeping analyte molecules in the proximity of the plasmonic metallic nanoparticles or nanostructures (George 2020; George et al. 2018; Barkur and Chidangil 2019; Xia et al. 2019; Hasna et al. 2016). The high sensitivity of the SERS technique is originated from the contribution of both electromagnetic and chemical enhancement mechanisms (Fig. 4). The concept of SERS, enhancement mechanisms, various fabrication methodologies employed for SERS substrate preparation and applications has been recently described (George 2020).
Illustration of the signal enhancement process in SERS (adapted from George (2020))
At present, a wide range of methodologies and protocols are being adopted to fabricate the SERS substrate, ranging from colloidal assisted approaches to advanced lithographic techniques (Shiohara et al. 2014). The plasmonic colloidal particle assisted SERS substrate fabrication technique relies on self-assembly or aggregation of the plasmonic particles, together with analyte molecules. The junction between the particle acts as hot spots to create large localized electromagnetic field to facilitate improved Raman scattering. Such approaches are reportedly able to detect analyte samples even at single molecule level (Shiohara et al. 2014). By varying the shape of the particles, the hot spot arrangement and consequently plasmonic field assisted Raman scattering signal enhancement can be varied to achieve improved signal strength. For example, anisotropic particles (e.g., star shaped or rod shaped) create higher field strength at the sharp edges of the corners and provide better signal strength for the molecules adhered there (Shiohara et al. 2014).
Gold nanorods are one of the most preferred structures since its optical properties can be highly tuned over a broad wavelength range based on its shape. Modifying the length to diameter (aspect ratio) of these nanoparticles can facilitate changes in surface plasmon band, which helps to utilize it for diverse applications. Another popular nanoparticle shape employed in SERS studies is the star-shaped particles (e.g., AuNS). The spikes of the NSs serve as an effective nanoantenna, which results in obtaining high electromagnetic field enhancement at the edge of each tip, leading to the creation of multiple hot spots within an individual nanoparticle. Owing to such improvements in Raman scattering of Ag nanoparticles, Ag nanostructures of different shapes (such as triangles, rods, and spheres) are employed in the SERS studies (Shiohara et al. 2014). Of these, the highest SERS enhancement is observed for star-shaped particles as compared to rod shaped and spherical Ag NPs due to localized electromagnetic field enhancement at hotspot between corners. Additionally, the spherical Ag NPs have the resonance band located at 400 nm whereas gold nanotriangles and nanostars have been found with multiple plasmon modes with a strong plane dipole band that allow resonance excitation of the Raman bands (Rycenga et al. 2011). Immobilization of the NPs on the substrate support material using linker molecules is a common practice (Toderas et al. 2007; Su et al. 2011). It is pertinent to note that Raman scattering is inversely proportional to the distance between the analyte and hot spot as I ~ 1/D12 (Ambartsumyan et al. 2020). Therefore, the proximity of the hotspot and analyte molecules are of paramount important in SERS studies. The SERS signal of the analyte reduces to zero at 3 nm from the hot spots (Dieringer et al. 2006; Masango et al. 2016). In the case of nanostructure-assisted SERS studies, the position of hotspots is fixed and thus the interface and molecule–substrate interaction have a pronounced effect on the SERS signal enhancement. The potential of anisotropic Ag nanoparticles (Ag NRs) has already been demonstrated to detect various viruses like adenovirus, human immunodeficiency virus, rhinovirus, and pathogens. Through integration of the improved stability of silver nanoparticles and enhancement factor of Ag nanoparticles, Au/Ag multilayered NRs arrays have been employed for the influenza A virus detection at very low concentrations (Ambartsumyan et al. 2020). The detection of viruses by the SERS technique can be made even more sensitive and specific by using reporter molecules (indirect detection method) attached with a sandwich immunoassay (Demirel et al. 2018). This route consists of a SERS tag, made up of a Raman reporter molecule and the recognition element, which is a specific antibody (detection antibody) confined on SERS-active nanoparticles substrate, and a template, called the capture substrate, which is functionalized with a linker antibody (capture antibody) to bind the antigen–SERS tag complex. Quantification is done by tracking the Raman signal from the reporter molecule before and after the SERS tag interacts with the capture element (Savinon-Flores et al. 2021) (Demirel et al. 2018).
Due to its flexibility, ease of use, and low cost, lateral flow immuno assay (LFIA) strips have been regarded as a suitable choice for POC diagnostics (Khlebtsov and Khlebtsov 2020). These sensors typically rely on visual readout avoiding any requirement for optical or electronic readout components. Efforts have been made to incorporate SERS tags with LFIA strips to improve the otherwise low sensitivity and limited quantitative/qualitative detection associated with conventional LFIA technique (Khlebtsov and Khlebtsov 2020). Kim et al. provided an in-depth review of Raman scattering-based lateral flow assay for infectious disease with reference to point of care applications. They have shown the efficacy of SERS multiplexed with lateral flow assay system in comparison with lateral flow immune assay (Kim et al. 2021b). Using SERS nanotags instead of solely gold nanoparticles can considerably enhance the Raman signals at hotspots, which can resolve the sensitivity issues in the standard LFIA detection. Use of Fe3O4/Ag magnetic SERS tags within the LFIA strips has facilitated an improved detection limit of 50 and 10 PFU/mL (plaque-forming units/mL) for influenza A H1N1 virus and human adenovirus (HAdV), respectively (Wang et al. 2019). In a similar work, multi-branched gold nanostars have been employed as SERS tags to provide a limit of detection (LOD) of 6.7 ng/mL for influenza A nucleoprotein, which shows a ~ 300-fold higher sensitivity as compared to conventional LFIA strips (Maneeprakorn et al. 2016). Hamad-Schifferli et al. developed a novel SERS-LFIA technique for the differentiation of the Zika virus and dengue virus nonstructural protein 1 (NS1) biomarkers (Sánchez-Purrà et al. 2017). That sensor employed SERS-encoded Au nanostars which resulted in the improvement of LOD ~ 7 and ~ 15 fold for dengue and Zika virus, respectively. Jeon et al. have developed a SERS-LFIA immunoassay, which can resolve the thermal stability concerns in the normal LFIA strips for their application in tropical regions. Using silica-encapsulated gold nanoparticles as tags for viral detection, the sensing strips were stable and reproducible at ~ 45 °C, whereas the solely gold nanoparticle-based LFIA strips suffered sensitivity issues (Jeon et al. 2020). Tang et al. reported a novel core–shell structure material, AuAg4 − ATP/AgNPs, as a Raman probe for the detection of avian influenza A (H7N9) virus with an estimated LOD of 0.0018 HAU (hemagglutination units) (Xiao et al. 2019). Similarly, Au-core Ag-shell nanoparticles were used for the detection of rotavirus in fecal samples using a SERS immunochromatographic assay (ICA) (Zhang et al. 2021c) with this assay able to quantitatively detect the virus in the range of 8–40,000 pg/mL with an LOD obtained as 8 pg/mL. Specific viral targets can be recognized by oligonucleotide aptamers and because of this the technique is gathering more interest recently (Kim et al. 2010). The popularity of the oligonucleotide-based SERS technique can be attributed to the ease of synthesis and purification process in addition to the potential for tagging and easy conjugation with metal and carbon-based nanostructures (Kim et al. 2010; Muhammad & Huang 2021). Moreover, the chaotic orientation, signal interference with SERS signal, and low thermal stability of antibodies compared with aptamers are promoting aptamer-based SERS platforms for virus detection (Tadesse et al. 2020; Tort et al. 2012).
Comparative studies using the SERS technique for investigation of different viruses
Recently, Amin Hadi et al. reviewed the applications of SERS in detection of biomaterials with special emphasis on COVID-19 detection. The article covers a vast spectra of details including the basic working principal of SERS to the proof of concept study demonstrating the applications in virus diagnostics (Eskandari et al. 2022). Yadav et al. proposed the use of a silver nanorod array, fabricated via the glancing angle deposition technique, for detection of the human immunodeficiency virus (HIV). Using a handheld SERS platform, this method was capable of selectively detecting HIV-1 subtypes in virus spiked blood plasma with minimal sample volume requirement (5 µL) (Yadav et al. 2021). A schematic diagram of the viral detection protocol using this SERS-based biosensor is shown as Fig. 5. SERS nanoprobes with gold shell-isolated nanoparticles were fabricated and conjugated with Nile blue dye used as the Raman reporter to achieve detection of Zika virus antigen at a concentration as low as 10 ng/mL. Further to this, the overlap of the Nile blue absorption band with the Raman probe laser excitation at ~ 633 nm can produce surface-enhanced resonance Raman scattering (SERRS), to enable accurate antigen detection (Camacho et al. 2018). Ryes et al. have developed a protein aggregation SERS detection scheme for detection of enterovirus from protein-rich samples (Reyes et al. 2017). In that study, Raman peak intensities at 510, 670, and 910 cm−1, which are dependent on the aggregation state of a specific gold nanoparticle-based bioconjugate, were analyzed to detect the presence of enterovirus 71. Others have demonstrated non-lithographically prepared metal/polymer nanostructured SERS substrates, which can carry out the detection of respiratory syncytial virus and coxsackievirus (Demirel et al. 2018). Antisense oligonucleotides (ASO) having complementary sequences with unique sites on viral genomes were considered as promising recognition elements in the Raman spectroscopic studies of viruses (Ambartsumyan et al. 2020) (Fig. 6).
Fabrication and working principle ((a)–(h)) of rapid handheld SERS platform for HIV detection (adapted from Yadav et al. (2021))
Schematic diagram demonstrating SERS detection of influenza virus (adapted from Chen et al. (2020))
The synthesis of ASO is relatively straightforward, and it includes the sequencing of the target virus genomes and the selection of a unique sequence for a specific viral strain of interest (Schubert and Kurreck 2006; Lenartowicz et al. 2016). Negri et al. have used an array of DNA oligonucleotides functionalized on a silver-based nanorod substrate for detecting generic markers linked with high pathogenicity in the influenza virus (Negri and Dluhy 2013). Another example demonstrating the potential of viral detection by SERS aptamer probes using the recently reported SERS imaging-based assays is illustrated in Fig. 6 (Chen et al. 2020). In this case, Chen et al. fabricated a 3D nano-popcorn substrate and functionalized it with thiolated captured DNAs for hybridizing Cy3-labeled aptamer probes for detecting influenza virus. The coupling between the virus and the aptamer probes resulted in the reduction of Raman signal intensity (Fig. 6C), which has been exploited for the quantitative detection of the virus. The possibility of influenza virus detection was also proposed by measuring the SERS signal from the aptamer-nucleoprotein complex generated after the binding of the anti-influenza aptamer to viral nucleoprotein (Negri et al. 2012).
SERS spectra of healthy and hepatitis C infected serum samples with different viral load (VL) (adapted from Kashif et al. (2020))
The SERS spectral signatures of the aptamer-nucleoprotein complex were able to effectively discriminate the selective binding of the aptamer-nucleoprotein from that of the aptamer alone. The use of gold-coated paramagnetic nanoparticles was reported as effective SERS substrates for targeting the DNAs derived from virus genomes (Zhang et al. 2012). Liu et al. have also employed graphene oxide-gold nanorods composite for the detection of hepatitis B surface antigen in serum with a limit of detection ~ \(0.05\;\mathrm{pg}\cdot\mathrm{mL}^{-1}\) (Liu et al. 2018). Lee et al. have reported a study demonstrating the use of SERS substrates for the detection of the HIV-1 within 5 s. Highly ordered Au nanodots deposited electrochemically on to indium tin oxide (ITO) substrates were able to detect HIV down to 35 fg/mL (Lee et al. 2015). In 2006, the literature reported a study using silver nanorod array substrates which was capable of rapid, differential detection of respiratory viruses and viral strains (Shanmukh et al. 2006). Two years later, the same group has reported SERS combined with chemometric tools for the reliable discrimination of closely related respiratory syncytial virus (RSV) strains and extended the work for human rotavirus detection and classification based on their genotypes (Shanmukh et al. 2008; Driskell et al. 2010). Lim et al. have also used SERS for influenza virus detection by exploiting enhanced spectral markers of their unique surface proteins/lipids generated with the aid of gold nanoparticle substrates. The technique was capable of differentiating non-influenza viruses from the influenza virus, and in addition, discrimination among different sub-types of influenza viruses. Determination of the presence of viral RNA mutations is important from both the preventative and therapeutic perspectives (Lim et al. 2015). The potential of the SERS technique to probe RNA mutations in the influenza A virus genome was shown using thiolated DNA hairpin functionalized gold nanostars (Dardir et al. 2020). In this case, a Raman active fluorescent dye was attached to 3’ terminus of a DNA sequence. The DNA hairpin structure was extended or folded in the presence or absence of viral RNA targets, respectively, causing the fluorophore to be close to the nanoparticle surface creating a kind of “OFF–ON” switching of the Raman signals. The detection was found to be sequence-specific, and the target detection at a single particle level was demonstrated. In addition, the signal recovery was found to be in correlation with the number of genetic mutations. Conventional cell lysing, RNA extraction, and preamplification procedures can be avoided in this SERS-based approach, which altogether yield a considerable reduction in detection cost and time (Dardir et al. 2020).
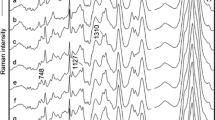
Gahlaut et al. have used a handheld Raman instrument, which enabled the detection of nonstructural protein (NS1) features from serum samples. Detection of NS1 protein, which is a secretory component of the virus, was utilized for the dengue diagnosis from serum (Gahlaut et al. 2020). Another recent work has demonstrated the possibility of hepatitis B viral load estimation and quantification from PCR products. Gel documentation systems (GDS) usually employed for the analysis of PCR products involve costly instrumentation, reagents, and lengthy UV exposure which can adversely impact the fluorescence of the bands. Batool et al. have analyzed the PCR product of viral RNA extracted from the hepatitis B patients using the SERS technique and the use of multivariate analytical tools such as principle component analysis (PCA), partial least squares-discriminant analysis (PLS-DA), and partial least squares regression (PLSR) resulted in a measured sensitivity of 89% and specificity of 98% (Batool et al. 2021). SERS measurements performed using silver nanoparticle solution on serum samples collected from hepatitis B virus-infected patients have shown an enhancement in the L-arginine Raman line at 493 cm−1 and a reduction in the nucleic acid bases in comparison with healthy volunteers (Lu et al. 2018). In addition, the peak assigned to saccharide at 889 cm−1 was found to be increased whereas the band of valine at 960 cm−1 decreased in diseased patients. This has been attributed to the saccharide metabolism disorder and the inhibition of the process of glycolysis (Lu et al. 2018). Additionally, markers for nonstructural protein (NS1) detection were identified from human saliva, which can be useful for monitoring viral infections (Othman et al. 2017). Recent studies have reported that SERS is capable for the detection of hepatitis C as well as quantification of viral load (VL) from blood serum samples (Nasir et al. 2021; Kashif et al. 2020). The peak present at 785 cm−1 originating due to the uracil base has been identified as a major marker for viral RNA, which is absent in normal serum samples, as evident from the spectra shown in Fig. 7. The increase in viral load has also resulted in the decrease in lipid contents seen in bands attributed to the lipid stretching at 1135 cm−1 and 1255 cm−1 (Fig. 8). The increase in RNA/DNA content in the clinical sample is in accordance with viral load measured from the Raman band at 1098 cm−1.
A Raman spectra of respiratory virus. B PCA plot of Raman spectral data (adapted from Yeh et al. (2020))
Microfluidic platforms are associated with benefits that include low sample volume, programmable sample handling, and automatic assay procedures (Bai et al. 2020). Incorporating the SERS technique into microfluidic platforms can pave way for the generation of automatic high-throughput on-chip detection (Bai et al. 2020; He et al. 2020; Ma et al. 2021; Burtsev et al. 2021; Gao et al. 2019). Recently, a microfluidic Raman platform that enables viral detection from clinical samples within a few minutes has been developed. The handheld device named “Virrion” allows for the capture of viruses depending on their size by use of a carbon nanotube forest arrays doped with gold nanoparticles patterned in a microfluidic channel, with later viral recognition from the patient sample executed using SERS followed by machine learning algorithms (Yeh et al. 2020). Raman spectra of respiratory viruses recorded using Virrion are shown in Fig. 8A and the PCA results (Fig. 8B) of the data were found to be effective in differentiating the viral strains. Hepatitis B virus antigen in blood plasma down to a level of 0.01 IU/mL was detected in a microfluidic system using SERS immunoassay (Kamińska et al. 2015). In that study, Kaminska et al. utilized a fuchsin-labeled immune-gold nanoflower together with SERS substrate for carrying out quantitative investigation on the virus with high sensitivity and selectivity. An Au–Ag coated GaN substrate embedded in the polycarbonate microfluidic device served as a SERS active substrate in this work (Kamińska et al. 2015). In a related study, Wang et al. have integrated digital microfluidics (DM) technology with the SERS technique for carrying out the automated and quantitative detection of avian influenza virus H5N1 in human serum (Wang et al. 2018). In that work, microfluidic chip was connected to a driving device, which can control the transport of a sample droplet to and from the original reservoirs upon application of an electric field to an array of electrodes on the substrate. The DM-SERS technique has shown a detection limit of 74 pg/mL with a shorter assay time (< 1 h) and low reagent consumption (~ 30 µL) with respect to conventional ELISA technique. A recent study has shown the ability of SERS technique to detect drug-resistant influenza virus in human nasopharyngeal aspirate using antibody mediated assays (Kim et al. 2021a).
Characterization of different viruses can be done via SERS by using specific reporter molecules attached to the immunoassay (Sivashanmugan et al. 2013). Viruses bind to a specific antibody functionalized on the nanostructures and hot spot formation happens. Characterization and quantification are done by tracking SERS signals from the reporter molecule before and after the SERS reporter molecule bind with the target analytes. Therefore, finding a particular Raman band or peak for each type of virus is not straightforward. In one study, Au/Ag multilayered nanorod arrays were prepared through the focused ion beam technique for the detection of the influenza A virus strain. The most intense peak for H2N2 was observed at 725 cm−1 (adenine), whereas the bands of adenine and tyrosine at 854 and 730 cm−1 were found to be prominent in the case of H3N2 (Sivashanmugan et al. 2013). Similarly, the intensity of the signal located at 1643 cm−1 in the case of A (H1N1) was more prominent. It has been observed that there are similarities in signal response between the SARS-CoV-2 virus and SARS-CoV-2 spike protein (YeonáYi & HyunáChung 2016). In a SERS study of different strains of the rotavirus, each virus strain generated similar SERS spectra with the major bands appearing at 1003, 1030, 1045, and 1592 cm−1. However, the SERS spectral fingerprints could differentiate samples as rotavirus-negative or rotavirus-positive. Using partial least squares regression (PLSR) analysis, SERS spectra for eight different rotavirus strains were able to classify each according to G and P genotype with an accuracy of more than 96% (Driskell et al. 2010). The details about the SERS substrates, the virus which was studied, and their corresponding Raman bands were given in Table 1.
SARS-CoV-2 detection
Many research groups have exploited Raman spectroscopy-based modalities for the detection of SARS-CoV-2 (Yang et al. 2021; Pramanik et al. 2021). Owing to its low scattering cross-section, detection of the SARS-CoV-2 virus is preferentially carried out using the SERS technique as compared to the traditional Raman spectroscopic technique. A SERS biosensor for detection of viruses was demonstrated using an ACE2-functionalized structured gold nanoneedle array (GNA) as “virus traps” (Yang et al. 2021). Using this technique, four proteins, namely, SARS-CoV-2 S protein, SARS-CoV-2 nucleocapsid protein, SARS-CoV S protein, and human ACE2 protein, could be characterized by measuring corresponding SERS spectra. The SERS spectra of those four proteins exhibited three characteristic peaks of SARS-CoV-2 S at 568 (amide V), 884 (trp), and 1296 cm−1 (amide III α helix) were apparent in all the recorded spectra. However, statistical analysis of the recorded Raman spectra using the PCA technique showed the potential of the technique to differentiate between individual spike proteins, namely, SARS-CoV-2 and SARS-CoV. Strains of the SARS-CoV-2 virus were analyzed in terms of their spike protein and nucleocapsid proteins, commonly represented as VS and VN, respectively (Yang et al. 2021). It was reported that the spike protein features exhibited at 568, 644, 884, 950, 1027, 1310, and 1445 cm−1 of the SERS spectra of SARS-COV-2 virus suggesting that the spike proteins are the major contributors to the Raman signature of the virus (Yang et al. 2021). Comparative studies of the inactivated and live SARS-CoV-2 virus exhibited substantial differences in spectral features, especially at 645, 884, 950, 1027, 1215, and 1445 cm−1 largely associated with the spike proteins. SERS signals of spike protein collected from a region of an area of 20 × 14 μm2 exhibited a relative standard deviation (RSD) for the Raman bands intensity (569 cm−1 and 1027 cm−1) of only 10.7% and 4.9%, indicating a high reproducibility. Using this technique binding between the ACE2 and SARS-CoV-2S is reported to be much more efficient than that between ACE2 and SARS-CoV-S. In addition, the study also elucidated that the viral infection process for SARS-CoV-2 is faster in comparison with the SARS-CoV version (Yang et al. 2021).
In a study based on colorimetric change, Au nanoparticles were functionalized with anti-spike antibody and used for rapid identification of SARS-CoV-2 antigen/virus within 5 min (Pramanik et al. 2021). Exhibiting rapidity, high selectivity, and high sensitivity, the SERS technique was used with 4-aminothiophenol as a reporter molecule adhered to the GNPs via an Au–S bond. When GNPs were added to the antigen or virus particles, antigen–antibody binding occurs which leads to the GNP aggregation and consequent color variation from pink to blue (Pramanik et al. 2021). Color change was observed by eye over a range bounded by a minimum concentration of 1 ng of COVID-19 antigen and 1000 virus particles per mL. SERS-based pseudo-SARS-CoV-2 virus detection using the anti-spike antibody and 4-aminothiophenol linked GNPs relies on the fact that in the presence of virus, the nanoparticles accumulate on the virus surface due to spike protein-antibody interaction. As the size of the virus is in the order of 120–150 nm, many functionalized gold nanoparticles can aggregate over each virus particle and form several hotspots and thus contribute to improved SERS signal. This interaction process took a maximum of 5 min to achieve the desired result and the Raman peak at 1078 cm−1 of anti-spike antibody and 4-aminothiophenol-functionalized gold nanoparticles was taken as reference. The intensity of the SERS peak at 1078 cm−1 varied linearly with the concentration of virus particles of pseudo-SARS-CoV-2. A LOD of 18 virus particles of SARS-CoV-2/mL could be detected, which is better than the present gold standard reverse transcription–polymerase chain reaction (RT-PCR) test where the LOD is 40 virus particle/mL (Pramanik et al. 2021).
A Raman spectroscopy investigation of SARS-CoV-2 in 17 water samples using a portable Raman spectrometer demonstrated the on-site measurement capability of the technique, in addition to the performance evaluation of the disinfection as well as viral survival in an environmental sample (Zhang et al. 2021a). That study employed the ACE2 receptor functionalized on silver nanorod array for the detection of viral presence in water (Liu et al. 2021). The binding of receptor-binding domain (RBD) of SARS-CoV-2 spike protein on the sensor array resulted in the quenching of most of the spectral features of the ACE2 receptor in addition to a shift in the band from 1189 to 1182 cm−1. The study has been suggested as a potential alternative to monitor the viral presence in wastewater treatment plants and pipe networks (Zhang et al. 2021a).
Even though LFIA methods have been reported for SARS-CoV-2 detection, the colorimetric analysis suffers shortcomings in quantitation and sensitivity (as discussed earlier). To resolve these concerns, Liu et al. demonstrated a fast and highly sensitive SERS-LFIA sensor for identifying SARS-CoV-2 antibodies in human serum with the experimental observations are summarized in Fig. 9 (Liu et al. 2021). They developed novel SERS tags by coating a complete Ag shell over an SiO2 core (SiO2/Ag) and conjugated this with viral spike protein for sensing purpose. The biosensor has been effective in the simultaneous detection of anti-SARS-CoV-2 IgG and IgM with excellent sensitivity as shown in Fig. 9 (as much as 800 times more sensitive than the conventional gold nanoparticle-based LFIA). The sensor performance was verified on 68 clinical samples with a demonstrated 100% accuracy and specificity of the SERS-LFIA. This technique can be advantageous for the screening of early infections, where the presence of antibodies in serum will be considerably lower.
IgM/IgG detection using A SERS-LFIA strips and B Au NP-based LFIA strips. C Raman spectra obtained for IgM and IgG lines. D Raman calibration plot obtained for IgM and IgG considering the band at 1328 cm.−1 (adapted from Liu et al. (2021))
Semiconductor-based SERS substrates have been utilized for ultra-sensitive detection of SARS-COV-2 protein (Peng et al. 2021). In this study, Ta2C MXene substrates have provided excellent SERS performance through photo-induced charge transfer (PICT) resonance enhancement and electromagnetic enhancement. This study demonstrated detection of SARS-CoV-2 spike protein, with a detection limit achieved as low as 5 × 10−9 M (Fig. 10). Somewhat similarly, Zhang et al. demonstrated a triple mode biosensing (colorimetric/SERS/fluorescence) approach for detection of SARS-CoV-2 RNA at femtomolar level within 40 min. The integration of multimodal approaches in biosensing can possibly increase the detection sensitivity and accuracy with reduction in false positive cases (Gao et al. 2021).
a Raman spectra of spike protein recorded with the different excitation wavelengths. b S-protein Raman spectra recorded using Ta2C NSs and Au NPS substrates (adapted from Peng et al. (2021))
Zhang et al. have demonstrated the possibility of SARS-CoV-2 spike protein detection from unprocessed saliva using SERS biosensing (Zhang et al. 2021b). SARS-CoV-2 spike antibodies conjugated to the SERS substrate fabricated via unique oil/water/oil three-phase liquid–liquid interfaces self-assembly process enabled ultrasensitive detection of SARS-CoV-2 spike protein n phosphate-buffered saline (0.77 fg mL−1) and untreated saliva (6.07 fg mL−1). The spectra obtained for different antigen concentrations in saliva have been shown in Fig. 11.
D SERS spectra obtained for various SARS-CoV-2 spike protein concentrations in saliva. E Comparison of Raman spectrum of spike protein in saliva and the blank signal (adapted from Zhang et al. (2021b))
Stanborough et al. have fabricated an aptamer-based SERS sensor for SARS-CoV-2 antigen protein detection at subpicomolar concentrations (Stanborough et al. 2021). The silver nanoparticle surface-functionalized with thiolated aptamer has shown depletion of free N–H stretching band upon the formation of strong amide hydrogen bonds during the selective, specific binding to spike protein. Another study involving aptamer receptors has been reported by Chen et al. where DNA aptamers immobilized on gold nano-popcorn surface were employed for sensing spike protein within 15 min (Chen et al. 2021). Detection limit ~ 10 PFU/mL was obtained for this study performed using SARS-CoV-2 lysate spiked in clinical samples, a comparatively better sensitivity with respect to commercial immunoassay rapid kit (~ 300 PFU/mL). Quantitative detection of SARS-CoV-2 virus along with high selectivity from other respiratory viruses via SERS was also demonstrated using aptamer-based silver colloidal solutions (Zavyalova et al. 2021). Huang et al. explored the diagnosis of SARS-CoV-2 antigen from clinical samples within 20 min using the deep learning-based SERS approach (Huang et al. 2021). This analytical approach utilized the hot spots generated by an aggregated spherical gold nanoparticle array for targeting the S protein of SARS-CoV-2. Upon the combination of a residual neural network (RNN)-based deep learning model, the study performed using a portable Raman spectrometer has shown promising results from throat swabs with sensitivity, specificity, and accuracy of 83.3%, 92.5%, and 87.7%, respectively, highlighting its potential as an onsite, rapid screening tool. The possibility of integrating the whole device inside a vehicle and thus the development of a mobile platform for field-based viral detection using this SERS approach was proposed in this work.
Goulart et al. have employed the Raman technique for the diagnosis of COVID-19 by identifying the spectral features of immunoglobulins IgM and IgG present in human serum (Goulart et al. 2021). Raman measurements have shown an increase in the characteristic markers attributed to nucleic acids, tryptophan, and immunoglobulins for COVID-19 cases. The study carried out on 54 control and 40 COVID-19 clinical samples was able to discriminate positive cases from control with a sensitivity and specificity of 84.0% and 95.0%, respectively, upon PLS-DA analysis. Carlomagno et al. have conducted SERS investigations on saliva collected from SARS-CoV-2 infected patients (COV +), healthy persons (CTRL), and patients with COVID-19 history (COV −) (Carlomagno et al. 2021). The spectral patterns obtained from three different classes are given in Fig. 12. The peaks present at 1048 and 1126 cm−1 have been found higher in patients infected with the virus and also in patients infected in the past with respect to control samples. The enhancement in these Raman peaks is indicative of aromatic amino acids (tryptophan and phenylalanine) abundant environment, which have been identified in the literature as one of the characteristic features of viruses of the coronavirus family. The spectral features obtained from saliva were sufficient enough for the discrimination of COVID-19 patients with accuracy, sensitivity, and specificity above 95%.
Raman spectra of healthy subjects (CTRL), subjects with viral infection (COV +), and subjects with history of viral infection (COV −) (adapted from Carlomagno et al. (2021))
In recent work, El-Said et al. have performed spectro-electrochemical detection of SARS-CoV-2 spike protein spiked in human serum samples (El-Said et al. 2022). Raman signal enhancement realized via the use of gold nanoparticles attached to a reduced porous graphene oxide modified substrate has resulted in obtaining a detection limit of 75 fmol L−1. A comparative evaluation of SARS-CoV-2 virions and purified RNA via Raman spectroscopy was reported in a recent proof-of-concept study, where a specific RNA marker band has been identified at 807 cm−1 (Wood et al. 2021). The potential of receptor binding domain (RBD) to be used as an effective target for SERS detection of the SARS-CoV-2 was also demonstrated recently (Awada et al. 2021). Ag/Au nanostructure fabricated on silicon rods via electroless etching and sputtering techniques was capable of detecting RBD protein up to 1 pM concentration in a short span (~ 3 s). In addition to detection, Raman spectroscopic tool has been also used to investigate the temperature response of spike protein in SARS-CoV-2 (Hernández-Arteaga et al. 2022). The spectral features of spike glycoprotein S1 started to show a significant intensity decline at 133 °C, whereas all the features were completely absent at 144 °C. SERS-based CRISPR (S-CRISPR) diagnostic test has been recently developed for detecting SARS-CoV-2 derived nucleic acids in RNA extracts collected from clinical specimens. This combined technique provides a reliable route for developing nucleic acid-based rapid point of care platforms, by eliminating the RNA amplification requirement in the traditional CRISPR/Cas-based tests. The various steps involved in viral identification from the nasopharyngeal swab samples were shown in Fig. 13 (Liang et al. 2021). The initial process comprised of the conventional RNA extraction from swab samples and further analysis was performed using a portable Raman plate reader. The presence of target DNA activates the Cas12a collateral cleavage, which separates the Raman tags from magnetic beads yielded a reduction in Raman signal intensity as shown in Fig. 13. In a recently published work, Choo et al. demonstrated a sensitive SERS technique for the detection of SARS-CoV-2 within 15 min using aptamer-based receptor on gold nano-popcorn substrate (Chen et al. 2021). Maiti et al. have demonstrated a proof of concept model for non-invasive COVID-19 detection by identifying the Raman signatures of saliva collected from infected patients at various stages. Using multivariate analytical tools with the developed Raman database, the model have been found effective in distinguishing healthy samples as well as different stages of disease recovery (Karunakaran et al. 2022). In another interesting work, the efficiency of COVID-19 vaccine has been verified from tear samples using SERS technique by synthesizing gold nanorods, nanoflowers, and nanospheres (Kim et al. 2022). The study conducted on limited sample size has been monitoring the spectral signatures of antibodies for identifying the vaccine status. The impact of the various nanostructure morphology on SERS-based viral detection have been investigated using bulk, dendritic, globular, and spiky silver nanostructures and found that dendrites have been showing efficacy in measurements in terms of sensitivity and repeatability (Li et al. 2022). Viral detection using SERS have been also reported by monitoring the changes in volatile organic compounds present in the exhaled breath samples within few minutes (Leong et al. 2022). In a recent work, SERS technique have been employed to evaluate the metabolic status in patients post COVID diagnosis (Chisanga et al. 2023). The study conducted in serum samples with a limited sample size (30 nos) has found variations in spectral signatures attributed to lipids and amino acids as possible markers in order to evaluate the disease recovery. Pack et al. have explored the high affinity of aptamers towards spike proteins for SERS-based detection with an LOD of 100 fg/mL (Park et al. 2023). The study employed aptamers coupled on a silver-based nanoforest surface prepared by sputtering technique for detecting recombinant spike protein by monitoring the Raman variations after protein-aptamer complex formation. Chiang et al. have developed a paper-based SERS substrate which can facilitate detection of SARS-CoV-2 variants which includes omicron, delta, and alpha (Yeh et al. 2023). The microplasma-engineered nanoassemblies fabricated on cellulose paper substrate were effective in enhancing the Raman signatures of spike proteins appearing at 2895 and 2946 cm−1.
Raman detection via collateral cleavage of SERS probe (adapted from Liang et al. (2021))
Raman spectroscopy-based technologies reported for SARS-CoV-2 by various research groups are given in Table 2.
Prospects and challenges
This work has reviewed the potential of various forms of Raman spectroscopy in the diagnosis of viruses such as SARS-CoV-2. The narrow peak emission bands of the Raman spectra facilitate the multiplexed detection of virus biomarkers. The lack of interference from water absorption in Raman spectroscopy (as compared to standard infrared spectroscopy) makes it a favored technology for the analysis of body fluids. Utilization of plasmonic nanostructures to enhance the molecular fingerprinting capability of Raman spectroscopy provides advantages related to a lower LOD, broader linear range, higher sensitivity, and greater selectivity in viral diagnosis. Functionalization of plasmonic nanoparticles/nanostructures facilitates the use of target-specific or biomarker-specific SERS technique which is a prerequisite in clinical settings for viral studies. The SERS technique also lessens the requirement of sample volume and allows for simultaneous targeting of a variety of biomarkers.
The ability of SERS for virus detection is dictated by factors such as substrate design, substrate biocompatibility, laser condition, and analyte characteristics. Gold nanoparticles exhibit greater stability compared to silver or copper nanoparticles; however, the ability of silver to induce strong LSPR makes it a potential candidate for SERS substrate fabrication. The trade-off between stability and induction of LSPR upon electromagnetic wave incidence decides the optimum substrate for harvesting the SERS signal (Sitjar et al. 2021). Other 2D materials, such as graphene, have also been investigated as SERS substrates and indeed the application of unique multi-layered SERS substrate—(Cu2O-Au)-graphene–Au—for effective sensing applications has been suggested in recent literature. These proposed structures have been shown capable of providing high field enhancement (of the order of ~ 108) at 1064 nm near-IR excitation, which can overcome problems related to visible light-induced fluorescence interference, photo-bleaching, and plasmonic heating, all of which impose limitations on the SERS effect (Nair & Murukeshan 2020). The orientation of analyte on the substrate surface also influences the SERS signal and hence has to be controlled by restricting the shape and size of nanoparticles. In the case of nanocavities, those virus particles having a size less than or comparable to the cavity can provide enhanced Raman signal. Xiao has come up with a “volume-enhanced Raman scattering (VERS)” approach which involves the fabrication of a substrate with volumetric nanoscale surface “hot spots.” The proposed substrate is made up of forming hollow nanocones at the bottom of microbowls (HNCMB). Unlike the conventional substrates, these VERS substrates facilitate an increase in the virus surface area which can enter the hotspots resulting in an enhanced collection of spectral information. These molecular imprinting-inspired substrates show superior single virus detection and reproducibility (Zhang et al. 2019). Virus particles attached to the top edges of nanorods can provide stronger signals since the major SERS effect is attributed to electromagnetic enhancement and the electromagnetic fields are stronger at sharp edges (Zhao et al. 2020). Spectral reproducibility is hampered by the non-uniform distribution and random aggregation of nanoparticles (Lu et al. 2022). The proper functionalization of affinity agents, such as antibodies and aptamers, on SERS substrates, can improve the specificity factor. Factors such as steric exclusion, random orientation on the substrate surface, and spectral interference tend to make aptamers a better candidate for functionalization of SERS substrate than antibodies (Tadesse et al. 2020).
The excitation wavelength and laser power play a critical role in the extraction of SERS signal with virus as analyte. Laser power considerations need to be balanced between extraction of a strong Raman signal without affecting substrate stability. Laser wavelength matching at (or near to) the plasmon resonance wavelength is desirable for the optimum SERS effect (Sitjar et al. 2021). In the resonance Raman spectroscopy technique, the laser wavelength is tuned to match with the electronic transition state of the molecule of interest but this requirement often demands a UV laser source for biomolecules. Virus integrity dictates the spectral features since the live virus, inactivated virus, enveloped virus, and non-enveloped virus are unique in their composition and activity. Mutations occurring in S-proteins (and other viral proteins) require regular updating of the SERS database. The investigation of clinical samples demands the rectification of Raman signature from other viruses present in the specimen, and so therefore multiplexed signal detection is mandatory. Advances in artificial intelligence and machine learning programs (that include multivariate statistical tools) allow for identification of receptor binding to viral spike proteins from SERS spectra. In view of the above, the setup for acquiring Raman signal from virus has to be designed to optimize signal strength by the considering the synergic effect all these factors.
The spatial resolution limitation associated with the traditional SERS approach can now be minimized by the use of TERS—which relies on the hot spot created at the apex of the SPM tip. Research on fabricating the TERS active sensor is still in progress, particularly with regard to the investigation of viruses. The TERS technique is more suitable for studying the mechanism of invasion/infection of viruses rather than their detection in patient samples. However, the effective utilization of the TERS technique demands significant user knowledge which currently limits the practical on-site applications via unskilled technicians, especially during pandemic crises (like COVID-19). Moreover, the associated low biological signal necessitates the fabrication of active probes that provide maximum plasmonic signal enhancement (Kumar et al. 2015). An alternate strategy involves using a higher integration time to achieve better signal strength; however, this often excludes recording the dynamics at a shorter duration of viral attack. Furthermore, the proximity of tips to the sample can lead to contamination or sample adsorption of the probe tip resulting in spurious results (Schmid et al. 2009). Though advances in micro and nanofabrication technologies have facilitated TERS tip fabrication, the fabrication of reproducible TERS tips is still a challenging task. Commonly adopted silver-based tips that provide high electric field enhancement are prone to oxidation or reaction with atmospheric sulfur consequently leading to deterioration of the plasmonic properties (Barrios et al. 2008). In addition, non-uniformity in sample surface features (e.g., roughness) can lead to fluctuating sample-probe distances and thus a varying TERS signal strength (Schmid et al. 2013).
In conclusion, although several features (such as rapid sampling, low LOD, excellent sensitivity and specificity, broad linear range) mark the SERS technique as particularly promising, its application in POC sensors for early viral detection is still at the beginning stages. The future potential of bifunctional nanoprobes for generating multiple spectral information (both Raman and fluorescence) from the complex biological samples should be explored further to enhance the sensitivity and specificity of virus detection (Xia et al. 2019). With the expectation of the integration of the SERS technique onto low-cost substrates like paper and polymers, the Raman spectroscopy technique is expected to provide fast, reliable, portable, low-cost sensors for the viral detection. Incorporation of the SERS technique into microfluidic platforms termed as “optofluidics” will facilitate high throughput sensing along with minimal sample volume requirement. Such highly reproducible, multiplexed, and molecule-specific SERS enhancement may enable a paradigm change in the POC technology for rapid detection of biomarkers from body fluids potentially becoming the preferred solution for rapid viral diagnosis in pandemics like COVID-19.
Data availability
Not applicable (as this is a review article there is no raw data available).
Code availability
Not applicable (as this is a review article there is no coding has been used).
References
Ali S, Hassan M, Saleem M, Tahir SF (2021) Deep transfer learning based hepatitis B virus diagnosis using spectroscopic images. Int J Imaging Syst Technol 31:94–105. https://doi.org/10.1002/ima.22462
Ambartsumyan O, Gribanyov D, Kukushkin V, Kopylov A, Zavyalova E (2020) SERS-based biosensors for virus determination with oligonucleotides as recognition elements. Int J Mol 21:3373. https://doi.org/10.3390/ijms21093373
Awada C, Abdullah MMB, Traboulsi H, Dab C, Alshoaibi A (2021) SARS-CoV-2 receptor binding domain as a stable-potential target for SARS-CoV-2 detection by surface—enhanced Raman spectroscopy. Sensors 21:4617. https://doi.org/10.3390/s21134617
Bai S, Serien D, Ma Y, Obata K, Sugioka K (2020) Attomolar sensing based on liquid interface-assisted surface-enhanced Raman scattering in microfluidic chip by femtosecond laser processing. ACS Appl Mater Interfaces 12:42328–42338. https://doi.org/10.1021/acsami.0c11322
Bankapur A, Barkur S, Chidangil S, Mathur D (2014) A micro-Raman study of live, single red blood cells (RBCs) treated with AgNO3 nanoparticles. PLoS One 9:e103493. https://doi.org/10.1371/journal.pone.0103493
Bankapur A, Zachariah E, Chidangil S, Valiathan M, Mathur D (2010) Raman tweezers spectroscopy of live, single red and white blood cells. PLoS One 5:e10427. https://doi.org/10.1371/journal.pone.0010427
Barkur S, Bankapur A, Pradhan M, Chidangil S, Mathur D, Ladiwala U (2015) Probing differentiation in cancer cell lines by single-cell micro-Raman spectroscopy. J Biomed Opt 20:085001. https://doi.org/10.1117/1.JBO.20.8.085001
Barkur S, Chidangil S (2019) Surface-enhanced Raman spectroscopy study of red blood cells and platelets. J Biomol Struct Dyn 37:1090–1098. https://doi.org/10.1080/07391102.2018.1450165
Barrios CA, Malkovskiy AV, Hartschuh RD, Kisliuk AM, Sokolov AP, Foster MD (2008) Extending lifetime of plasmonic silver structures designed for high-resolution chemical imaging or chemical and biological sensing. Proc Chem, Biol, Radiol, Nuclear, Explosives (CBRNE) sensing IX, SPIE, 63–74.https://doi.org/10.1117/12.777304
Batool F, Nawaz H, Majeed MI et al (2021) SERS-based viral load quantification of hepatitis B virus from PCR products. Spectrochim Acta Part A: Mol Biomol Spectrosc 255:119722. https://doi.org/10.1016/j.saa.2021.119722
Burrell CJ, Howard CR, Murphy FA (2017) Laboratory diagnosis of virus diseases. Fenner White’s Med Virol 135. https://doi.org/10.1016/B978-0-12-375156-0.00010-2
Burtsev V, Erzina M, Guselnikova O et al (2021) Detection of trace amounts of insoluble pharmaceuticals in water by extraction and SERS measurements in a microfluidic flow regime. Analyst 146:3686–3696. https://doi.org/10.1039/D0AN02360D
Butler HJ, Ashton L, Bird B et al (2016) Using Raman spectroscopy to characterize biological materials. Nat Protoc 11:664–687. https://doi.org/10.1038/nprot.2016.036
Camacho SA, Sobral-Filho RG, Aoki PHB, Constantino CJL, Brolo AG (2018) Zika immunoassay based on surface-enhanced Raman scattering nanoprobes. ACS Sensors 3:587–594. https://doi.org/10.1021/acssensors.7b00639
Carlomagno C, Bertazioli D, Gualerzi A et al (2021) COVID-19 salivary Raman fingerprint: innovative approach for the detection of current and past SARS-CoV-2 infections. Sci Rep 11:4943. https://doi.org/10.1038/s41598-021-84565-3
Chen H, Park S-G, Choi N et al (2021) Sensitive detection of SARS-CoV-2 using a SERS-based aptasensor. ACS Sensors 6:2378–2385. https://doi.org/10.1021/acssensors.1c00596
Chen H, Park S-G, Choi N et al (2020) SERS imaging-based aptasensor for ultrasensitive and reproducible detection of influenza virus A. Biosens Bioelectron 167:112496. https://doi.org/10.1016/j.bios.2020.112496
Chisanga M, Williams H, Boudreau D, Pelletier JN, Trottier S, Masson J-F (2023) Label-free SERS for rapid differentiation of SARS-CoV-2-induced serum metabolic profiles in non-hospitalized adults. Anal Chem. https://doi.org/10.1021/acs.analchem.2c04514
Cialla D, Deckert-Gaudig T, Budich C et al (2009) Raman to the limit: tip-enhanced Raman spectroscopic investigations of a single tobacco mosaic virus. J Raman Spectrosc: Int J Original Work All Aspects Raman Spectrosc Including Higher Order Processes, Also Brillouin Rayleigh Scattering 40:240–243. https://doi.org/10.1002/jrs.2123
Culha M, Cullum B, Lavrik N, Klutse CK (2012) Surface-enhanced Raman scattering as an emerging characterization and detection technique. J Nanotechnol 2012.https://doi.org/10.1155/2012/971380
Dardir K, Wang H, Martin BE, Atzampou M, Brooke CB, Fabris L (2020) SERS nanoprobe for intracellular monitoring of viral mutations. J Phys Chem C 124:3211–3217. https://doi.org/10.1021/acs.jpcc.9b09253
Das RS, Agrawal Y (2011) Raman spectroscopy: recent advancements, techniques and applications. Vib Spectrosc 57:163–176. https://doi.org/10.1016/j.vibspec.2011.08.003
Deckert V, Deckert-Gaudig T, Cialla-May D, et al. (2020) Laser spectroscopic technique for direct identification of a single virus I: FASTER CARS. Proc Natl Acad Sci 117, 27820-27824https://doi.org/10.1073/pnas.201316911
Demirel G, Usta H, Yilmaz M, Celik M, Alidagi HA, Buyukserin F (2018) Surface-enhanced Raman spectroscopy (SERS): an adventure from plasmonic metals to organic semiconductors as SERS platforms. J Mater Chem C 6:5314–5335. https://doi.org/10.1039/C8TC01168K
Desai S, Mishra SV, Joshi A et al (2020) Raman spectroscopy-based detection of RNA viruses in saliva: a preliminary report. J Biophotonics 13:e202000189. https://doi.org/10.1002/jbio.202000189
Dieringer JA, Mcfarland AD, Shah NC et al (2006) Introductory lecture surface enhanced Raman spectroscopy: new materials, concepts, characterization tools, and applications. Faraday Discuss 132:9–26. https://doi.org/10.1039/B513431P
Dietzek B, Cialla D, Schmitt M, Popp J (2010) Introduction to the fundamentals of Raman spectroscopy. In. Confocal Raman microscopy. Springer, 21–42. https://doi.org/10.1007/978-3-642-12522-5_2
Dou T, Li Z, Zhang J, Evilevitch A, Kurouski D (2020) Nanoscale structural characterization of individual viral particles using atomic force microscopy infrared spectroscopy (AFM-IR) and tip-enhanced Raman spectroscopy (TERS). Anal Chem 92:11297–11304. https://doi.org/10.1021/acs.analchem.0c01971
Driskell JD, Zhu Y, Kirkwood CD, Zhao Y, Dluhy RA, Tripp RA (2010) Rapid and sensitive detection of rotavirus molecular signatures using surface enhanced Raman spectroscopy. PLoS One 5:e10222. https://doi.org/10.1371/journal.pone.0010222
El-Said WA, Al-Bogami AS, Alshitari W (2022) Synthesis of gold nanoparticles@ reduced porous graphene-modified ITO electrode for spectroelectrochemical detection of SARS-CoV-2 spike protein. Spectrochim Acta Part A: Mol Biomol Spectrosc 264:120237. https://doi.org/10.1016/j.saa.2021.120237
Eskandari V, Sahbafar H, Zeinalizad L, Hadi A (2022) A review of applications of surface-enhanced Raman spectroscopy laser for detection of biomaterials and a quick glance into its advances for COVID-19 investigations. ISSS J Micro Smart Syst 11:363–382. https://doi.org/10.1007/s41683-022-00103-x
Espy M, Uhl J, Sloan L et al (2006) Real-time PCR in clinical microbiology: applications for routine laboratory testing. Clin Microbiol Rev 19:165–256. https://doi.org/10.1128/CMR.19.1.165-256.2006
Fan C, Hu Z, Riley LK, Purdy GA, Mustapha A, Lin M (2010) Detecting food-and waterborne viruses by surface-enhanced Raman spectroscopy. J Food Sci 75:M302–M307. https://doi.org/10.1111/j.1750-3841.2010.01619.x
Gahlaut S, Savargaonkar D, Sharan C, Yadav S, Mishra P, Singh J (2020) SERS platform for dengue diagnosis from clinical samples employing a hand held Raman spectrometer. Anal Chem 92:2527–2534. https://doi.org/10.1021/acs.analchem.9b04129
Gao R, Lv Z, Mao Y et al (2019) SERS-based pump-free microfluidic chip for highly sensitive immunoassay of prostate-specific antigen biomarkers. ACS Sensors 4:938–943. https://doi.org/10.1021/acssensors.9b00039
Gao Y, Han Y, Wang C et al (2021) Rapid and sensitive triple-mode detection of causative SARS-CoV-2 virus specific genes through interaction between genes and nanoparticles. Anal Chim Acta 1154:338330. https://doi.org/10.1016/j.aca.2021.338330
George JE, Unnikrishnan V, Mathur D, Chidangil S, George SD (2018) Flexible superhydrophobic SERS substrates fabricated by in situ reduction of Ag on femtosecond laser-written hierarchical surfaces. Sens Actuators, B Chem 272:485–493. https://doi.org/10.1016/j.snb.2018.05.155
George SD (2020) Surface-enhanced Raman scattering substrates: fabrication, properties, and applications. Self-standing Substrates: Mater Appl 83–118. https://doi.org/10.1007/978-3-030-29522-6_3
Gogone IC, Ferreira GH, Gava D et al (2021) Applicability of Raman spectroscopy on porcine parvovirus and porcine circovirus type 2 detection. Spectrochim Acta Part A: Mol Biomol Spectrosc 249:119336. https://doi.org/10.1016/j.saa.2020.119336
Goulart ACC, Zângaro RA, Carvalho HC, Silveira L Jr (2021) Diagnosing COVID-19 in human sera with detected immunoglobulins IgM and IgG by means of Raman spectroscopy. J Raman Spectrosc 52:2671–2682. https://doi.org/10.1002/jrs.6235
Hamden KE, Bryan BA, Ford PW, Xie C, Li Y-Q, Akula SM (2005) Spectroscopic analysis of Kaposi’s sarcoma-associated herpesvirus infected cells by Raman tweezers. J Virol Methods 129:145–151. https://doi.org/10.1016/j.jviromet.2005.05.018
Hasna K, Lakshmi K, Ezhuthachan Jayaraj MK, Kumar KR, Matham MV (2016) Development of high-sensitive, reproducible colloidal surface-enhanced Raman spectroscopy active substrate using silver nanocubes for potential biosensing applications. J Nanophotonics 10:026020. https://doi.org/10.1117/1.JNP.10.026020
He X, Ge C, Zheng X et al (2020) Rapid identification of alpha-fetoprotein in serum by a microfluidic SERS chip integrated with Ag/Au nanocomposites. Sens Actuators B: Chem 317:128196. https://doi.org/10.1016/j.snb.2020.128196
Hematian A, Sadeghifard N, Mohebi R et al (2016) Traditional and modern cell culture in virus diagnosis. Osong Public Health Res Perspect 7:77–82. https://doi.org/10.1016/j.phrp.2015.11.011
Hermann P, Hermelink A, Lausch V et al (2011) Evaluation of tip-enhanced Raman spectroscopy for characterizing different virus strains. Analyst 136:1148–1152. https://doi.org/10.1039/C0AN00531B
Hernández-Arteaga A, Ojeda-Galván H, Rodríguez-Aranda M et al (2022) Determination of the denaturation temperature of the spike protein S1 of SARS-CoV-2 (2019 nCoV) by Raman spectroscopy. Spectrochim Acta Part A: Mol Biomol Spectrosc 264:120269. https://doi.org/10.1016/j.saa.2021.120269
Huang J, Wen J, Zhou M et al (2021) On-site detection of SARS-CoV-2 antigen by deep learning-based surface-enhanced Raman spectroscopy and its biochemical foundations. Anal Chem 93:9174–9182. https://doi.org/10.1021/acs.analchem.1c01061
Huang T-X, Huang S-C, Li M-H, Zeng Z-C, Wang X, Ren B (2015) Tip-enhanced Raman spectroscopy: tip-related issues. Anal Bioanal Chem 407:8177–8195. https://doi.org/10.1007/s00216-015-8968-8
Ibrahim O, Toner M, Flint S, Byrne HJ, Lyng FM (2021) The potential of Raman spectroscopy in the diagnosis of dysplastic and malignant oral lesions. Cancers 13:619. https://doi.org/10.3390/cancers13040619
Jeon J, Lee SH, Joung Y, Kim K, Choi N, Choo J (2020) Improvement of reproducibility and thermal stability of surface-enhanced Raman scattering-based lateral flow assay strips using silica-encapsulated gold nanoparticles. Sens Actuators B: Chem 321:128521. https://doi.org/10.1016/j.snb.2020.128521
Kamińska A, Witkowska E, Winkler K, Dzięcielewski I, Weyher JL, Waluk J (2015) Detection of hepatitis B virus antigen from human blood: SERS immunoassay in a microfluidic system. Biosens Bioelectron 66:461–467. https://doi.org/10.1016/j.bios.2014.10.082
Karunakaran V, Joseph MM, Yadev I et al (2022) A non-invasive ultrasensitive diagnostic approach for COVID-19 infection using salivary label-free SERS fingerprinting and artificial intelligence. J Photochem Photobiol B: Biol 234:112545. https://doi.org/10.1016/j.jphotobiol.2022.112545
Kashif M, Majeed MI, Hanif MA, Ur Rehman A (2020) Surface enhanced Raman spectroscopy of the serum samples for the diagnosis of hepatitis C and prediction of the viral loads. Spectrochim Acta Part A: Mol Biomol Spectrosc 242:118729. https://doi.org/10.1016/j.saa.2020.118729
Khlebtsov B, Khlebtsov N (2020) Surface-enhanced raman scattering-based lateral-flow immunoassay. Nanomaterials 10:2228. https://doi.org/10.3390/nano10112228
Kim H, Kang H, Kim H-N et al (2021) Development of 6E3 antibody-mediated SERS immunoassay for drug-resistant influenza virus. Biosens Bioelectron 187:113324. https://doi.org/10.1016/j.bios.2021.113324
Kim K, Kashefi-Kheyrabadi L, Joung Y et al (2021) Recent advances in sensitive surface-enhanced Raman scattering-based lateral flow assay platforms for point-of-care diagnostics of infectious diseases. Sens Actuators B: Chem 329:129214. https://doi.org/10.1016/j.snb.2020.129214
Kim NH, Lee SJ, Moskovits M (2010) Aptamer-mediated surface-enhanced Raman spectroscopy intensity amplification. Nano Lett 10:4181–4185. https://doi.org/10.1021/nl102495j
Kim W, Kim S, Han J et al (2022) An excitation wavelength-optimized, stable SERS biosensing nanoplatform for analyzing adenoviral and AstraZeneca COVID-19 vaccination efficacy status using tear samples of vaccinated individuals. Biosens Bioelectron 204:114079. https://doi.org/10.1016/j.bios.2022.114079
Kumar N, Mignuzzi S, Su W, Roy D (2015) Tip-enhanced Raman spectroscopy: principles and applications. EPJ Techniques and Instrumentation 2:1–23. https://doi.org/10.1140/epjti/s40485-015-0019-5
Kuzmin AN, Pliss A, Lim C-K et al (2016) Resonance Raman probes for organelle-specific labeling in live cells. Sci Rep 6:28483. https://doi.org/10.1038/srep28483
Lee J-H, Kim B-C, Byeung-Keun O, Choi J-W (2015) Rapid and sensitive determination of HIV-1 virus based on surface enhanced Raman spectroscopy. J Biomed Nanotechnol 11:2223–2230. https://doi.org/10.1166/jbn.2015.2117
Lenartowicz E, Nogales A, Kierzek E, Kierzek R, Martínez-Sobrido L, Turner DH (2016) Antisense oligonucleotides targeting influenza A segment 8 genomic RNA inhibit viral replication. Nucleic Acid Ther 26:277–85. https://doi.org/10.1089/nat.2016.0619
Leong SX, Leong YX, Tan EX et al (2022) Noninvasive and point-of-care surface-enhanced Raman scattering (SERS)-based breathalyzer for mass screening of coronavirus disease 2019 (COVID-19) under 5 min. ACS Nano 16:2629–2639. https://doi.org/10.1021/acsnano.1c09371
Li Z, Luo Y, Song Y, Zhu Q, Xu T, Zhang X (2022) One-click investigation of shape influence of silver nanostructures on SERS performance for sensitive detection of COVID-19. Anal Chim Acta 1234:340523. https://doi.org/10.1016/j.aca.2022.340523
Liang J, Teng P, Xiao W et al (2021) Application of the amplification-free SERS-based CRISPR/Cas12a platform in the identification of SARS-CoV-2 from clinical samples. J Nanobiotechnol 19:1–9. https://doi.org/10.1186/s12951-021-01021-0
Lim J-Y, Nam J-S, Shin H et al (2019) Identification of newly emerging influenza viruses by detecting the virally infected cells based on surface enhanced Raman spectroscopy and principal component analysis. Anal Chem 91:5677–5684. https://doi.org/10.1021/acs.analchem.8b05533
Lim J-Y, Nam J-S, Yang S-E et al (2015) Identification of newly emerging influenza viruses by surface-enhanced Raman spectroscopy. Anal Chem 87:11652–11659. https://doi.org/10.1021/acs.analchem.5b02661
Liu H, Dai E, Xiao R et al (2021) Development of a SERS-based lateral flow immunoassay for rapid and ultra-sensitive detection of anti-SARS-CoV-2 IgM/IgG in clinical samples. Sens Actuators B: Chem 329:129196. https://doi.org/10.1016/j.snb.2020.129196
Liu M, Zheng C, Cui M et al (2018) Graphene oxide wrapped with gold nanorods as a tag in a SERS based immunoassay for the hepatitis B surface antigen. Microchim Acta 185:1–8. https://doi.org/10.1007/s00604-018-2989-x
Lu Y, Lin L, Ye J (2022) Human metabolite detection by surface-enhanced Raman spectroscopy. Mater Today Bio 100205.https://doi.org/10.1016/j.mtbio.2022.100205
Lu Y, Lin Y, Zheng Z et al (2018) Label free hepatitis B detection based on serum derivative surface enhanced Raman spectroscopy combined with multivariate analysis. Biomed Opt Express 9:4755–4766. https://doi.org/10.1364/BOE.9.004755
Lukose J, Mithun N, Mohan G, Shastry S, Chidangil S (2019) Normal saline-induced deoxygenation of red blood cells probed by optical tweezers combined with the micro-Raman technique. RSC Adv 9:7878–7884. https://doi.org/10.1039/C8RA10061F
Ma L, Ye S, Wang X, Zhang J (2021) SERS-microfluidic approach for the quantitative detection of miRNA using DNAzyme-mediated reciprocal signal amplification. ACS Sensors 6:1392–1399. https://doi.org/10.1021/acssensors.1c00063
Maneeprakorn W, Bamrungsap S, Apiwat C, Wiriyachaiporn N (2016) Surface-enhanced Raman scattering based lateral flow immunochromatographic assay for sensitive influenza detection. RSC Adv 6:112079–112085. https://doi.org/10.1039/C6RA24418A
Masango SS, Hackler RA, Large N et al (2016) High-resolution distance dependence study of surface-enhanced Raman scattering enabled by atomic layer deposition. Nano Lett 16:4251–4259. https://doi.org/10.1021/acs.nanolett.6b01276
Monsees I, Turzynski V, Esser SP et al (2022) Label-free Raman microspectroscopy for identifying prokaryotic virocells. Msystems 7:e01505-e1521. https://doi.org/10.1128/msystems.01505-21
Moor K, Ohtani K, Myrzakozha D, Zhanserkenova O, Andriana BB, Sato H (2014) Noninvasive and label-free determination of virus infected cells by Raman spectroscopy. J Biomed Opt 19:067003. https://doi.org/10.1117/1.JBO.19.6.067003
Moor K, Terada Y, Taketani A, Matsuyoshi H, Ohtani K, Sato H (2018) Early detection of virus infection in live human cells using Raman spectroscopy. J Biomed Opt 23:097001. https://doi.org/10.1117/1.JBO.23.9.097001
Muhammad M, Huang Q (2021) A review of aptamer-based SERS biosensors: design strategies and applications. Talanta 227:122188. https://doi.org/10.1016/j.talanta.2021.122188
Nair RV, Murukeshan VM (2020) (Cu2O-Au)–graphene-Au layered structures as efficient near infra-red SERS substrates. Sci Rep 10:1–10. https://doi.org/10.1038/s41598-020-60874-x
Naseer K, Amin A, Saleem M, Qazi J (2019) Raman spectroscopy based differentiation of typhoid and dengue fever in infected human sera. Spectrochim Acta Part A Mol Biomol Spectrosc 206:197–201. https://doi.org/10.1016/j.saa.2018.08.008
Nasir S, Majeed MI, Nawaz H et al (2021) Surface enhanced Raman spectroscopy of RNA samples extracted from blood of hepatitis C patients for quantification of viral loads. Photodiagnosis Photodynamic Ther 33:102152. https://doi.org/10.1016/j.pdpdt.2020.102152
Negri P, Chen G, Kage A et al (2012) Direct optical detection of viral nucleoprotein binding to an anti-influenza aptamer. Anal Chem 84:5501–5508. https://doi.org/10.1021/ac202427e
Negri P, Dluhy RA (2013) Detection of genetic markers related to high pathogenicity in influenza by SERS. Analyst 138:4877–4884. https://doi.org/10.1039/C3AN00774J
Olschewski K, Kämmer E, Stöckel S et al (2015) A manual and an automatic TERS based virus discrimination. Nanoscale 7:4545–4552. https://doi.org/10.1039/C4NR07033J
Othman N, Lee KY, Radzol A, Mansor W, Ramlan N (2017) Linear discriminant analysis for detection of salivary NS1 from SERS spectra. Proc TENCON 2017–2017 IEEE Region 10 Conf IEEE 2876–2879.https://doi.org/10.1109/TENCON.2017.8228352
Park KS, Choi A, Kim HJ, et al. (2023) Ultra-sensitive label-free SERS biosensor with high-throughput screened DNA aptamer for universal detection of SARS-CoV-2 variants from clinical samples. Biosens Bioelectron 115202. https://doi.org/10.1016/j.bios.2023.115202
Peng Y, Lin C, Long L et al (2021) Charge-transfer resonance and electromagnetic enhancement synergistically enabling MXenes with excellent SERS sensitivity for SARS-CoV-2 S protein detection. Nano-Micro Letters 13:1–17. https://doi.org/10.1007/s40820-020-00565-4
Pilat Z, Jonas A, Pilatova J et al (2020) Analysis of bacteriophage–host interaction by Raman tweezers. Anal Chem 92:12304–12311. https://doi.org/10.1021/acs.analchem.0c01963
Popp J, Kiefer W (2006) Raman scattering, fundamentals. Encycl Anal Chem: Appl Theory Instrum. https://doi.org/10.1002/9780470027318.a6405
Pramanik A, Gao Y, Patibandla S et al (2021) The rapid diagnosis and effective inhibition of coronavirus using spike antibody attached gold nanoparticles. Nanoscale Adv 3:1588–1596. https://doi.org/10.1039/D0NA01007C
Ramoji A, Pahlow S, Pistiki A et al (2022) Understanding viruses and viral infections by biophotonic methods. Transl Biophotonics 4:e202100008. https://doi.org/10.1002/tbio.202100008
Rehman A, Anwar S, Firdous S, Ahmed M, Rasheed R, Nawaz M (2012) Dengue blood analysis by Raman spectroscopy. Laser Phys 22:1085–1089. https://doi.org/10.1134/S1054660X12060072
Reyes M, Piotrowski M, Ang SK et al (2017) Exploiting the anti-aggregation of gold nanostars for rapid detection of hand, foot, and mouth disease causing enterovirus 71 using surface-enhanced Raman spectroscopy. Anal Chem 89:5373–5381. https://doi.org/10.1021/acs.analchem.7b00066
Rybicki E (1990) The classification of organisms at the edge of life or problems with virus systematics. S A J Sci 86:182. https://hdl.handle.net/10520/AJA00382353_6229
Rycenga M, Cobley CM, Zeng J et al (2011) Controlling the synthesis and assembly of silver nanostructures for plasmonic applications. Chem Rev 111:3669–3712. https://doi.org/10.1021/cr100275d
Saade J, Pacheco MTT, Rodrigues MR, Silveira L Jr (2008) Identification of hepatitis C in human blood serum by near-infrared Raman spectroscopy. Spectroscopy 22:387–395. https://doi.org/10.3233/SPE-2008-0344
Saleem M, Bilal M, Anwar S, Rehman A, Ahmed M (2013) Optical diagnosis of dengue virus infection in human blood serum using Raman spectroscopy. Laser Phys Lett 10:035602. https://doi.org/10.1088/1612-2011/10/3/035602
Salman A, Shufan E, Zeiri L, Huleihel M (2014) Characterization and detection of Vero cells infected with herpes simplex virus type 1 using Raman spectroscopy and advanced statistical methods. Methods 68:364–370. https://doi.org/10.1016/j.ymeth.2014.02.022
Sánchez-Purrà M, Carré-Camps M, De Puig H, Bosch I, Gehrke L, Hamad-Schifferli K (2017) Surface-enhanced Raman spectroscopy-based sandwich immunoassays for multiplexed detection of Zika and dengue viral biomarkers. ACS Infectious Diseases 3:767–776. https://doi.org/10.1021/acsinfecdis.7b00110
Sanchez JE, Jaramillo SA, Settles E et al (2021) Detection of SARS-CoV-2 and its S and N proteins using surface enhanced Raman spectroscopy. RSC Adv 11:25788–25794. https://doi.org/10.1021/acsinfecdis.7b00110
Savinon-Flores F, Mendez E, Lopez-Castanos M et al (2021) A review on SERS-based detection of human virus infections: influenza and coronavirus. Biosensors 11:66. https://doi.org/10.3390/bios11030066
Schmid T, Opilik L, Blum C, Zenobi R (2013) Nanoscale chemical imaging using tip-enhanced Raman spectroscopy: a critical review. Angew Chem Int Ed 52:5940–5954. https://doi.org/10.1002/anie.201203849
Schmid T, Yeo BS, Leong G, Stadler J, Zenobi R (2009) Performing tip-enhanced Raman spectroscopy in liquids. J Raman Spectrosc 40:1392–1399. https://doi.org/10.1002/jrs.2387
Schubert S, Kurreck J, 2006. Oligonucleotide-based antiviral strategies. RNA Towards Med 261–87. https://doi.org/10.1007/3-540-27262-3_13
Shanmukh S, Jones L, Driskell J, Zhao Y, Dluhy R, Tripp RA (2006) Rapid and sensitive detection of respiratory virus molecular signatures using a silver nanorod array SERS substrate. Nano Lett 6:2630–2636. https://doi.org/10.1021/nl061666f
Shanmukh S, Jones L, Zhao Y-P, Driskell J, Tripp R, Dluhy R (2008) Identification and classification of respiratory syncytial virus (RSV) strains by surface-enhanced Raman spectroscopy and multivariate statistical techniques. Anal Bioanal Chem 390:1551–1555. https://doi.org/10.1007/s00216-008-1851-0
Shiohara A, Wang Y, Liz-Marzán LM (2014) Recent approaches toward creation of hot spots for SERS detection. J Photochem Photobiol, C 21:2–25. https://doi.org/10.1016/j.jphotochemrev.2014.09.001
Shipp DW, Sinjab F, Notingher I (2017) Raman spectroscopy: techniques and applications in the life sciences. Adv Opt Photon 9:315–428. https://doi.org/10.1364/AOP.9.000315
Sitjar J, Liao J-D, Lee H, Tsai H-P, Wang J-R, Liu P-Y (2021) Challenges of SERS technology as a non-nucleic acid or-antigen detection method for SARS-CoV-2 virus and its variants. Biosens Bioelectron 181:113153. https://doi.org/10.1016/j.bios.2021.113153
Sivashanmugan K, Liao J-D, You J-W, Wu C-L (2013) Focused-ion-beam-fabricated Au/Ag multilayered nanorod array as SERS-active substrate for virus strain detection. Sens Actuators, B Chem 181:361–367. https://doi.org/10.1016/j.snb.2013.01.035
Stanborough T, Given FM, Koch B et al (2021) Optical detection of CoV-SARS-2 viral proteins to sub-picomolar concentrations. ACS Omega 6:6404–6413. https://doi.org/10.1021/acsomega.1c00008
Storch GA (2000) Diagnostic virology. Clin Infect Dis 31:739–751. https://doi.org/10.1086/314015
Su Q, Ma X, Dong J, Jiang C, Qian W (2011) A reproducible SERS substrate based on electrostatically assisted APTES-functionalized surface-assembly of gold nanostars. ACS Appl Mater Interf 3:1873–1879. https://doi.org/10.1021/am200057f
Tadesse LF, Safir F, Ho C-S et al (2020) Toward rapid infectious disease diagnosis with advances in surface-enhanced Raman spectroscopy. J Chem Phys 152:240902. https://doi.org/10.1063/1.5142767
Tiwari D, Jakhmola S, Pathak DK, Kumar R, Jha HC (2020) Temporal in vitro Raman spectroscopy for monitoring replication kinetics of Epstein-Barr virus infection in glial cells. ACS Omega 5:29547–29560. https://doi.org/10.1021/acsomega.0c04525
Toderas F, Baia M, Baia L, Astilean S (2007) Controlling gold nanoparticle assemblies for efficient surface-enhanced Raman scattering and localized surface plasmon resonance sensors. Nanotechnology 18:255702. https://doi.org/10.1088/0957-4484/18/25/255702
Tort N, Salvador J-P, Avino A et al (2012) Synthesis of steroid–oligonucleotide conjugates for a DNA site-encoded SPR immunosensor. Bioconjug Chem 23:2183–2191. https://doi.org/10.1021/bc300138p
Wang C, Wang C, Wang X et al (2019) Magnetic SERS strip for sensitive and simultaneous detection of respiratory viruses. ACS Appl Mater Interf 11:19495–19505. https://doi.org/10.1021/acsami.9b03920
Wang Y, Ruan Q, Lei Z-C et al (2018) Highly sensitive and automated surface enhanced Raman scattering-based immunoassay for H5N1 detection with digital microfluidics. Anal Chem 90:5224–5231. https://doi.org/10.1021/acs.analchem.8b00002
Wen ZQ, Armstrong A, Thomas GJ (1999) Demonstration by ultraviolet resonance Raman spectroscopy of differences in DNA organization and interactions in filamentous viruses Pf1 and fd. Biochemistry 38:3148–3156. https://doi.org/10.1021/bi981965m
Wen ZQ, Overman SA, Thomas GJ (1997) Structure and interactions of the single-stranded DNA genome of filamentous virus fd: investigation by ultraviolet resonance Raman spectroscopy. Biochemistry 36:7810–7820. https://doi.org/10.1021/bi970342q
Wen ZQ, Thomas GJ Jr (1998) UV resonance Raman spectroscopy of DNA and protein constituents of viruses: assignments and cross sections for excitations at 257, 244, 238, and 229 nm. Biopolymers Original Res Biomol 45:247–256. https://doi.org/10.1002/(SICI)1097-0282(199803)45:3%3c247::AID-BIP7%3e3.0.CO;2-R
Wood BR, Kochan K, Bedolla DE et al (2021) Infrared based saliva screening test for COVID-19. Angew Chem 133:17239–17244. https://doi.org/10.1002/ange.202104453
Wood BR, Langford SJ, Cooke BM et al (2004) Resonance Raman spectroscopy reveals new insight into the electronic structure of β-hematin and malaria pigment. J Am Chem Soc 126:9233–9239. https://doi.org/10.1021/ja038691x
Wood BR, Mcnaughton D (2006) Resonance Raman spectroscopy in malaria research. Expert Rev Proteomics 3:525–544. https://doi.org/10.1586/14789450.3.5.525
Xia Y, Padmanabhan P, Sarangapani S, Gulyás B, Vadakke Matham M (2019) Bifunctional fluorescent/Raman nanoprobe for the early detection of amyloid. Sci Rep 9:1–11. https://doi.org/10.1038/s41598-019-43288-2
Xiao M, Xie K, Dong X et al (2019) Ultrasensitive detection of avian influenza A (H7N9) virus using surface-enhanced Raman scattering-based lateral flow immunoassay strips. Anal Chim Acta 1053:139–147. https://doi.org/10.1016/j.aca.2018.11.056
Yadav S, Senapati S, Desai D, Gahlaut S, Kulkarni S, Singh J (2021) Portable and sensitive Ag nanorods based SERS platform for rapid HIV-1 detection and tropism determination. Colloids Surfaces B: Biointerfaces 198:111477. https://doi.org/10.1016/j.colsurfb.2020.111477
Yang Y, Peng Y, Lin C et al (2021) Human ACE2-functionalized gold “virus-trap” nanostructures for accurate capture of SARS-CoV-2 and single-virus SERS detection. Nano-Micro Lett 13:1–13. https://doi.org/10.1007/s40820-021-00620-8
Yeh Y-J, Le T-N, Hsiao WW-W, Tung K-L, Ostrikov KK, Chiang W-H (2023) Plasmonic nanostructure-enhanced Raman scattering for detection of SARS-CoV-2 nucleocapsid protein and spike protein variants. Analy Chim Acta 1239:340651. https://doi.org/10.1016/j.aca.2022.340651
Yeh Y-T, Gulino K, Zhang Y, et al. (2020) A rapid and label-free platform for virus capture and identification from clinical samples. Proc Natl Acad Sci 117:895–901.https://doi.org/10.1073/pnas.191011311
Yeonáyi S, Hyunáchung B (2016) Facile and sensitive detection of influenza viruses using SERS antibody probes. RSC Adv 6:84415–84419. https://doi.org/10.1039/C6RA13966C
Zavyalova E, Ambartsumyan O, Zhdanov G et al (2021) SERS-based aptasensor for rapid quantitative detection of SARS-CoV-2. Nanomaterials 11:1394. https://doi.org/10.3390/nano11061394
Zhang D, Zhang X, Ma R et al (2021) Ultra-fast and onsite interrogation of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) in waters via surface enhanced Raman scattering (SERS). Water Res 200:117243. https://doi.org/10.1016/j.watres.2021.117243
Zhang H, Harpster MH, Wilson WC, Johnson PA (2012) Surface-enhanced Raman scattering detection of DNAs derived from virus genomes using Au-coated paramagnetic nanoparticles. Langmuir 28:4030–4037. https://doi.org/10.1021/la204890t
Zhang M, Li X, Pan J et al (2021) Ultrasensitive detection of SARS-CoV-2 spike protein in untreated saliva using SERS-based biosensor. Biosens Bioelectron 190:113421. https://doi.org/10.1016/j.bios.2021.113421
Zhang X, Zhang X, Luo C et al (2019) Volume-enhanced Raman scattering detection of viruses. Small 15:1805516. https://doi.org/10.1002/smll.201805516
Zhang Y, Wu G, Wei J et al (2021c) Rapid and sensitive detection of rotavirus by surface-enhanced Raman scattering immunochromatography. Microchim Acta 188:1–10. https://doi.org/10.1007/s00604-020-04670-4
Zhao X, Wen J, Zhu A et al (2020) Manipulation and applications of hotspots in nanostructured surfaces and thin films. Nanomaterials 10:1667. https://doi.org/10.3390/nano10091667
Zhao Y, Tian S, Yu L, Zhang Z, Zhang W (2021) Analysis and classification of hepatitis infections using Raman spectroscopy and multiscale convolutional neural networks. J Appl Spectrosc 88:441–451. https://doi.org/10.1007/s10812-021-01192-6
Zhu X, Xu T, Lin Q, Duan Y (2014) Technical development of Raman spectroscopy: from instrumental to advanced combined technologies. Appl Spectrosc Rev 49:64–82. https://doi.org/10.1080/05704928.2013.798801
Funding
Open access funding provided by Manipal Academy of Higher Education, Manipal. Mr Ajaya Kumar Barik, Mr Mithun N and Mr Sanoop Pavithran M are very thankful to Dr TMA Pai PhD Scholarships of Manipal Academy of Higher Education, Manipal.
Author information
Authors and Affiliations
Contributions
Review conception, manuscript preparation and editing were done by Jijo Lukose. Ajaya Kumar Barik, Mithun N, and Sanoop Pavithran M contributed to manuscript preparation. Sajan D. George and V.M. Murukeshan contributed to editing and structuring the manuscript. Review conception, and editing were done by Santhosh Chidangil. All authors commented on previous versions of the manuscript and all authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval
Not applicable.
Consent to participate
Not applicable.
Consent for publication
Not applicable.
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Lukose, J., Barik, A.K., Mithun N et al. Raman spectroscopy for viral diagnostics. Biophys Rev 15, 199–221 (2023). https://doi.org/10.1007/s12551-023-01059-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12551-023-01059-4