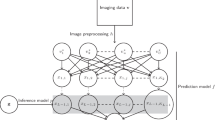
Abstract
Objective
To identify candidate neuroimaging and genetic biomarkers for Alzheimer’s disease (AD) and other brain disorders, especially for little-investigated brain diseases, we advocate a data-driven approach which incorporates an adaptive classifier ensemble model acquired by integrating Convolutional Neural Network (CNN) and Ensemble Learning (EL) with Genetic Algorithm (GA), i.e., the CNN-EL-GA method, into Genome-Wide Association Studies (GWAS).
Methods
Above all, a large number of CNN models as base classifiers were trained using coronal, sagittal, or transverse magnetic resonance imaging slices, respectively, and the CNN models with strong discriminability were then selected to build a single classifier ensemble with the GA for classifying AD, with the help of the CNN-EL-GA method. While the acquired classifier ensemble exhibited the highest generalization capability, the points of intersection were determined with the most discriminative coronal, sagittal, and transverse slices. Finally, we conducted GWAS on the genotype data and the phenotypes, i.e., the gray matter volumes of the top ten most discriminative brain regions, which contained the ten most points of intersection.
Results
Six genes of PCDH11X/Y, TPTE2, LOC107985902, MUC16 and LINC01621 as well as Single-Nucleotide Polymorphisms, e.g., rs36088804, rs34640393, rs2451078, rs10496214, rs17016520, rs2591597, rs9352767 and rs5941380, were identified.
Conclusion
This approach overcomes the limitations associated with the impact of subjective factors and dependence on prior knowledge while adaptively achieving more robust and effective candidate biomarkers in a data-driven way.
Significance
The approach is promising to facilitate discovering effective candidate genetic biomarkers for brain disorders, as well as to help improve the effectiveness of identified candidate neuroimaging biomarkers for brain diseases.





Similar content being viewed by others
References
Hong-meng L, Di Z, Xue-bin C (2017) Deep learning for early diagnosis of Alzheimer’s disease based on intensive alexnet. Comput Sci 44(6):50–59. https://doi.org/10.11896/j.issn.1002-137X.2017.6A.011
Prince M, Wimo A, Guerchet M, Ali G, Wu Y, Prina M (2015) World Alzheimer report 2015. the global impact of dementia. Alzheimer’s Disease International (ADI), London
Liu S, Liu S, Cai W, Pujol S, Kikinis R, Feng D (2014) Early diagnosis of Alzheimer’s disease with deep learning. Int Sympos Biomed Imaging. https://doi.org/10.1109/ISBI.2014.6868045
Frisoni GB, Fox NC, Jack CR, Scheltens P, Thompson PM (2010) The clinical use of structural MRI in Alzheimer disease. Nat Rev Neurol 6(2):67–77. https://doi.org/10.1038/nrneurol.2009.215
Hao X, Bao Y, Guo Y, Yu M, Zhang D, Risacher SL, Saykin AJ, Yao X, Shen L, Initiative ADN et al (2020) Multi-modal neuroimaging feature selection with consistent metric constraint for diagnosis of Alzheimer’s disease. Med Image Anal 60:101625. https://doi.org/10.1016/j.media.2019.101625
Jie B, Liu M, Liu J, Zhang D, Shen D (2016) Temporally constrained group sparse learning for longitudinal data analysis in Alzheimer’s disease. IEEE Trans Biomed Eng 64(1):238–249. https://doi.org/10.1109/tbme.2016.2553663
Liu M, Li F, Yan H, Wang K, Ma Y, Shen L, Xu M, Initiative ADN et al (2020) A multi-model deep convolutional neural network for automatic hippocampus segmentation and classification in Alzheimer’s disease. Neuroimage 208:116459. https://doi.org/10.1016/j.neuroimage.2019.116459
Liu M, Zhang D, Adeli E, Shen D (2015) Inherent structure-based multiview learning with multitemplate feature representation for Alzheimer’s disease diagnosis. IEEE Trans Biomed Eng 63(7):1473–1482. https://doi.org/10.1109/tbme.2015.2496233
Liu M, Zhang J, Adeli E, Shen D (2018) Joint classification and regression via deep multi-task multi-channel learning for Alzheimer’s disease diagnosis. IEEE Trans Biomed Eng 66(5):1195–1206. https://doi.org/10.1109/tbme.2018.2869989
Tong T, Gao Q, Guerrero R, Ledig C, Chen L, Rueckert D, Initiative ADN et al (2016) A novel grading biomarker for the prediction of conversion from mild cognitive impairment to Alzheimer’s disease. IEEE Trans Biomed Eng 64(1):155–165. https://doi.org/10.1109/TBME.2016.2549363
Zeng N, Qiu H, Wang Z, Liu W, Zhang H, Li Y (2018) A new switching-delayed-PSO-based optimized SVM algorithm for diagnosis of Alzheimer’s disease. Neurocomputing 320:195–202. https://doi.org/10.1016/j.neucom.2018.09.001
Zhou T, Thung KH, Liu M, Shen D (2018) Brain-wide genome-wide association study for Alzheimer’s disease via joint projection learning and sparse regression model. IEEE Trans Biomed Eng 66(1):165–175. https://doi.org/10.1109/tbme.2018.2824725
Rathore S, Habes M, Iftikhar MA, Shacklett A, Davatzikos C (2017) A review on neuroimaging-based classification studies and associated feature extraction methods for Alzheimer’s disease and its prodromal stages. Neuroimage 155:530–548. https://doi.org/10.1016/j.neuroimage.2017.03.057
LeCun Y, Bottou L, Bengio Y, Haffner P (1998) Gradient-based learning applied to document recognition. Proc IEEE 86(11):2278–2324. https://doi.org/10.1109/5.726791
Litjens G, Kooi T, Bejnordi BE, Setio AAA, Ciompi F, Ghafoorian M, Van Der Laak JA, Van Ginneken B, Sanchez CI (2017) A survey on deep learning in medical image analysis. Med Image Anal 42:60–88. https://doi.org/10.1016/j.media.2017.07.005
Shen D, Wu G, Suk HI (2017) Deep learning in medical image analysis. Annu Rev Biomed Eng 19:221–248. https://doi.org/10.1146/annurev-bioeng-071516-044442
Hosseini-Asl E, Keynton R, El-Baz A (2016) Alzheimer’s disease diagnostics by adaptation of 3d convolutional network. IEEE Int Conf Image Process. https://doi.org/10.1109/ICIP.2016.7532332
Jain R, Jain N, Aggarwal A, Hemanth DJ (2019) Convolutional neural network based Alzheimer’s disease classification from magnetic resonance brain images. Cogn Syst Res 57:147–159. https://doi.org/10.1016/j.cogsys.2018.12.015
Kruthika K, Maheshappa H, Initiative ADN et al (2019) Cbir system using capsule networks and 3d cnn for Alzheimer’s disease diagnosis. Inform Med Unlocked 14:59–68. https://doi.org/10.1016/j.imu.2019.100227
Tanveer M, Richhariya B, Khan R, Rashid A, Khanna P, Prasad M, Lin C (2020) Machine learning techniques for the diagnosis of Alzheimer’s disease: A review. ACM Trans Multimed Comput Commun Appl 16(1):1–35. https://doi.org/10.1145/3344998
Wang H, Shen Y, Wang S, Xiao T, Deng L, Wang X, Zhao X (2019) Ensemble of 3d densely connected convolutional network for diagnosis of mild cognitive impairment and Alzheimer’s disease. Neurocomputing 333:145–156. https://doi.org/10.1016/j.neucom.2018.12.018
Pan D, Zeng A, Jia L, Huang Y, Frizzell T, Song X (2020) Early detection of Alzheimer’s disease using magnetic resonance imaging: a novel approach combining convolutional neural networks and ensemble learning. Front Neurosci. https://doi.org/10.3389/fnins.2020.00259
Hariri AR, Weinberger DR (2003) Imaging genomics. Br Med Bull 65(1):259–270. https://doi.org/10.1002/9783527678679.dg05936
Chauhan G, Adams HH, Bis JC, Weinstein G, Yu L, Toglhofer AM, Smith AV, Van Der Lee SJ, Gottesman RF, Thomson R et al (2015) Association of alzheimer’s disease gwas loci with mri markers of brain aging. Neurobiol Aging 36(4):1765-e7. https://doi.org/10.1016/j.neurobiolaging.2014.12.028
Tan L, Yu JT, Zhang W, Wu ZC, Zhang Q, Liu QY, Wang W, Wang HF, Ma XY, Cui WZ (2013) Association of GWAS-linked loci with late-onset Alzheimer’s disease in a northern Han Chinese population. Alzheimer’s Dement 9(5):546–553. https://doi.org/10.1016/j.jalz.2012.08.007
Salvatore C, Cerasa A, Battista P, Gilardi MC, Quattrone A, Castiglioni I (2015) Magnetic resonance imaging biomarkers for the early diagnosis of Alzheimer’s disease: a machine learning approach. Front Neurosci 9:307. https://doi.org/10.3389/fnins.2015.00307
Chang CC, Chow CC, Tellier LC, Vattikuti S, Purcell SM, Lee JJ (2015) Second-generation PLINK: rising to the challenge of larger and richer datasets. GigaScience. https://doi.org/10.1186/s13742-015-0047-8
Fischl B (2012) FreeSurfer. Neuroimage 62(2):774–781. https://doi.org/10.1016/j.neuroimage.2012.01.021
Fan L, Li H, Zhuo J, Zhang Y, Wang J, Chen L, Yang Z, Chu C, Xie S, Laird AR, Fox PT, Eickhoff SB, Yu C, Jiang T (2016) The human brainnetome atlas: a new brain atlas based on connectional architecture. Cereb Cortex 26(8):3508–3526. https://doi.org/10.1093/cercor/bhw157
Wang SH, Phillips P, Sui Y, Liu B, Yang M, Cheng H (2018) Classification of Alzheimer’s disease based on eight-layer convolutional neural network with leaky rectified linear unit and max pooling. J Med Syst. https://doi.org/10.1007/s10916-018-0932-7
Sul JH, Martin LS, Eskin E (2018) Population structure in genetic studies: Confounding factors and mixed models. PLoS Genet 14(12):e1007309. https://doi.org/10.1371/journal.pgen.1007309
Liew SS, Khalil-Hani M, Bakhteri R (2016) Bounded activation functions for enhanced training stability of deep neural networks on visual pattern recognition problems. Neurocomputing 216:718–734. https://doi.org/10.1016/j.neucom.2016.08.037
Kingma DP, Ba J (2015) Adam: A method for stochastic optimization. In: Bengio Y, LeCun Y (eds) International Conference on Learning Representations, Conference Track Proceedings, ICLR 2015, San Diego, CA, USA, May 7–9, 2015
Pan D, Huang Y, Zeng A, Jia L, Song X, A.D.N.I. et al (2019) Early diagnosis of Alzheimer’s disease based on deep learning and GWAS. International workshop on human brain and artificial intelligence. Springer, Berlin, pp 52–68. https://doi.org/10.1007/978-981-15-1398-5_4
Carrasquillo MM, Zou F, Pankratz VS, Wilcox SL, Ma L, Walker LP, Younkin SG, Younkin CS, Younkin LH, Bisceglio GD et al (2009) Genetic variation in PCDH11X is associated with susceptibility to late-onset Alzheimer’s disease. Nat Genet 41(2):192–198. https://doi.org/10.1038/ng.305
Beecham GW, Naj A, Gilbert JR, Haines JL, Buxbaum JD, Pericak-Vance MA (2010) PCDH11X variation is not associated with late-onset Alzheimer disease susceptibility. Psychiatr Genet 20(6):321. https://doi.org/10.1097/YPG.0b013e32833b635d
Rosenberg RN, Lambracht-Washington D, Yu G, Xia W (2016) Genomics of Alzheimer disease: a review. JAMA Neurol 73(7):867–874. https://doi.org/10.1001/jamaneurol.2016.0301
Kantojarvi K (2013) Exploring genetic susceptibility to autism spectrum disorders. Academic Dissertation, University of Helsinki
Liu X, Cheng R, Verbitsky M, Kisselev S, Browne A, Mejia-Sanatana H, Louis ED, Cote LJ, Andrews H, Waters C et al (2011) Genome-wide association study identifies candidate genes for Parkinson’s disease in an Ashkenazi Jewish population. BMC Med Genet 12(1):104. https://doi.org/10.1186/1471-2350-12-104
Pankratz N, Wilk JB, Latourelle JC, DeStefano AL, Halter C, Pugh EW, Doheny KF, Gusella JF, Nichols WC, Foroud T, Richard HM (2008) Genomewide association study for susceptibility genes contributing to familial Parkinson disease. Hum Genet 124(6):593–605. https://doi.org/10.1007/s00439-008-0582-9
Galichon P, Mesnard L, Hertig A, Stengel B, Rondeau E (2012) Unrecognized sequence homologies may confound genome-wide association studies. Nucleic Acids Res 40(11):4774–4782. https://doi.org/10.1093/nar/gks169
Jean PS (2008) Genes associated with schizophrenia identified using a whole genome scan. US Patent App. 11/970,611
Staley LA (2018) Analysis of whole exome sequence data in affected cousin pairs from high-risk Alzheimer’s pedigrees. In: Master thesis, all Theses and Dissertations, Brigham Young University, p 7332
Bi XA, Liu Y, Xie Y, Hu X, Jiang Q (2020) Morbigenous brain region and gene detection with a genetically evolved random neural network cluster approach in late mild cognitive impairment. Bioinformatics 36(8):2561–2568. https://doi.org/10.1093/bioinformatics/btz967
Bi XA, Hu X, Wu H, Wang Y (2020) Multimodal data analysis of Alzheimer’s disease based on clustering evolutionary random forest. IEEE J Biomed Health Inform 24(10):2973–2983. https://doi.org/10.1109/jbhi.2020.2973324
Acknowledgements
We thank the reviewers for their constructive comments on this article. This study was supported by NSF of China (Grant Nos. 61976058, 61772143 and U19A2067), National Key RD Program of China (Grant No. 2018YFC0910405), Science and Technology Planning Project of Guangdong (Grant No. 2021A1515012300, 2019A050510041 and 2020A1515010941), and Science and Technology Planning Project of Guangzhou (Grant Nos. 202103000034 and 202002020090). Additional funding for neurological research was from Surrey Hospital & Outpatient Centre Foundation (FHG2017-001). Data collection and sharing for this project were funded by the Alzheimer’s Disease Neuroimaging Initiative (ADNI) (National Institutes of Health Grant U01 AG024904) and DOD ADNI (Department of Defense Award No. W81XWH-12-2-0012). ADNI was funded by the National Institute on Aging, the National Institute of Biomedical Imaging and Bioengineering, and through generous contributions from the following: AbbVie, Alzheimer’s Association; Alzheimer’s Drug Discovery Foundation; Araclon Biotech; BioClinica, Inc.; Biogen; Bristol-Myers Squibb Company; CereSpir, Inc.; Cogstate; Eisai Inc.; Elan Pharmaceuticals, Inc.; Eli Lilly and Company; EuroImmun; F. Hoffmann-La Roche Ltd and its affiliated company Genentech, Inc.; Fujirebio; GE Healthcare; IXICO Ltd.; Janssen Alzheimer Immunotherapy Research & Development, LLC.; Johnson & Johnson Pharmaceutical Research & Development LLC.; Lumosity; Lundbeck; Merck & Co., Inc.; Meso Scale Diagnostics, LLC.; NeuroRx Research; Neurotrack Technologies; Novartis Pharmaceuticals Corporation; Pfizer Inc.; Piramal Imaging; Servier; Takeda Pharmaceutical Company; and Transition Therapeutics. The Canadian Institutes of Health Research was providing funds to support ADNI clinical sites in Canada. Private sector contributions are facilitated by the Foundation for the National Institutes of Health (www.fnih.org). The grantee organization is the Northern California Institute for Research and Education, and the study is coordinated by the Alzheimer’s Therapeutic Research Institute at the University of Southern California. ADNI data are disseminated by the Laboratory for Neuro Imaging at the University of Southern California. Additional funding support for the manuscript preparation was from the Surrey Hospital & Outpatient Centre Foundation of Fraser Health, Canada.
Author information
Authors and Affiliations
Consortia
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Alzheimer’s Disease Neuroimaging Initiative-Data used in preparation of this article were obtained from the Alzheimer's Disease Neuroimaging Initiative (ADNI) database (adni.loni.usc.edu). As such, the investigators within the ADNI contributed to the design and implementation of ADNI and/or provided data but did not participate in analysis or writing of this report. A complete listing of ADNI investigators can be found at: http://adni.loni.usc.edu/wpcontent/uploads/how_to_apply/ADNI_Acknowledgement_List.pdf.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Zeng, A., Rong, H., Pan, D. et al. Discovery of Genetic Biomarkers for Alzheimer’s Disease Using Adaptive Convolutional Neural Networks Ensemble and Genome-Wide Association Studies. Interdiscip Sci Comput Life Sci 13, 787–800 (2021). https://doi.org/10.1007/s12539-021-00470-3
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12539-021-00470-3