Abstract
Background
Alzheimer’s disease (AD) is an important, progressive neurodegenerative disease, with a complex genetic architecture. A key goal of biomedical research is to seek out disease risk genes, and to elucidate the function of these risk genes in the development of disease. For this purpose, expanding the AD-associated gene set is necessary. In past research, the prediction methods for AD related genes has been limited in their exploration of the target genome regions. We here present a genome-wide method for AD candidate genes predictions.
Methods
We present a machine learning approach (SVM), based upon integrating gene expression data with human brain-specific gene network data, to discover the full spectrum of AD genes across the whole genome.
Results
We classified AD candidate genes with an accuracy and the area under the receiver operating characteristic (ROC) curve of 84.56% and 94%. Our approach provides a supplement for the spectrum of AD-associated genes extracted from more than 20,000 genes in a genome wide scale.
Conclusions
In this study, we have elucidated the whole-genome spectrum of AD, using a machine learning approach. Through this method, we expect for the candidate gene catalogue to provide a more comprehensive annotation of AD for researchers.
Similar content being viewed by others
Background
Alzheimer’s disease (AD) is a widespread progressive neurodegenerative disease type, characterized by impaired memory, cognitive functioning, and changed behavior [1]. Past genetic research implicates b-amyloid peptide accumulation and deposition, as well as tau protein pathology, selective neuronal death, synaptic and neurotransmitter loss, and neuroinflammation in Alzheimer’s disease pathogenesis [2]. However, the standard of research into AD is Genome Wide Association Studies (GWAS) with pedigree analysis, rather than candidate pathway exploration. Therefore, the understanding of AD is limited by sample size and quality, making it a challenge to have overall insight into AD. Moreover, AD heritability is estimated at ~60–80% [3], while the genetic architecture of AD is imperfectly characterized.
Complex human diseases such as AD are caused by the composite action of multiple, disease-related genes. At present, AD has at least 4 well-known disease-causing genes: the amyloid precursor protein (APP) gene and the Presenilin (PSEN1/PSEN2) genes for familial AD, and apolipoprotein E (APOE) ε4 for sporadic AD [1]. A key goal of biomedical research is to seek out disease risk genes, and to elucidate the function of these risk genes in the development of disease and the complex networks of gene-gene interactions underlying complex traits [4]. For this purpose, expanding the AD-associated gene set is necessary. However, with the rapid development of sequencing technology, large amounts of new sequence data must be analyzed to extract disease-related genes using novel, computational approaches.
Thus far, methods based on different data-types and different strategies have been applied in predicting AD-associated genes. Prediction methods can be roughly divided into five types: methods integrating protein-protein interaction networks with information such as protein subcellular localization, gene expression quantification, or gene functional annotation [5,6,7,8]; patterns of sequence-based features shared by disease genes [9,10,11]; machine learning and network topological features [12]; or information about tissue-specific networks [13, 14]. In past research, these methods have been applied to predict associated genes or biomarkers [15,16,17]. But there are few reports on the predictions based on the brain gene expression data.
We here present a genome-wide method using human brain-specific gene interaction network constructed by gene expression data, resulting in predictions for AD candidate genes. The brain-specific network works by integrating relations between each pair of AD-associated genes, in order to present how genes function together in the brain. This disease-gene classifier extracts the correlation coefficients of known AD-associated/AD-unassociated genes in this brain network, and then uses the coefficients specific to AD-associated/AD-unassociated genes to predict the level of potential AD association for every gene in the genome. After this initial prediction, we then select the predicted AD-related genes with GO functional annotations which are same as those of most of known AD-association genes as the candidate risk genes or biomarkers for AD. In addition, we compared the sequence-based features of all genes, in order to assess our approach. Furthermore, we found that some of our predictions are consistent with associations reported elsewhere in the AD literature, validating the result. The genome-wide complement of Alzheimer’s candidate genes predicted in this study can thus be used to explore the mechanism of AD, and ultimately, to assist in the discovery of better treatments for AD.
Methods
Data sources
AD related genes
We collected 335 AD-associated genes from public Alzheimer’s disease databases (AlzGene, http://www.alzgene.org/) and from publications treating upon AD. Then, we collected total 22,646 genes and removed the 335 AD-associated genes and genes recorded in OMIM (https://www.omim.org/) as our initial control dataset. Finally, we selected 335 AD non-associated genes (the same number of AD-associated genes) from the initial control dataset with the minimal interaction between 335 AD-associated genes (Optimal Control Dataset). At the same time, we randomly selected the other dataset of 335 non-associated genes (n = 100) for SVM training, but the Optimal Control Dataset had the highest correct rate (Additional file 1: Figure S1).
Gene-gene interaction data
The machine learning method used require a set of known gene-gene interaction data for the model input. We obtained this data from GIANT [18] (http://giant.princeton.edu), which can be set to extract the subset of tissue-specific interactions. Since the pathogenesis of Alzheimer’s disease is associated with brain tissue, we selected brain-specific, functional gene interaction data.
Prediction
There are many different supervised machine learning approaches. We implement a prediction algorithm by SVM (Support Vector Machines) using the e1071 package of R, published by David Meyer. SVM was chosen here, as it can allow the assignment of different weights to different classes. The kernel type used in training and predicting is radial. In addition, a 5-fold cross validation was performed, in order to assess the quality of the model.
Results
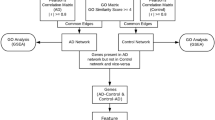
We used human, tissue-specific networking to discover the full spectrum of AD genes across the whole genome. We then assessed the reliability of these genes (Fig. 1). Here, we depict the AD genetic spectrum, and provide an overview of the AD associated genes, honing the focus of further AD study for future researchers.
Genome-wide prediction of Alzheimer-associated genes. Our prediction was based on machine learning methods that are trained upon the already associated AD genes, which are already linked to AD at various levels of evidence (C1AD-C4AD), as a positive training set, and other genes excluded from the OMIM database (C5) as a negative training set. Combining these with the human brain-specific gene network, we were able to build an evidence-weighted, network-based classifier, and predict the probability of the association between each gene and AD across the genome
Alzheimer’s disease genes spectrum
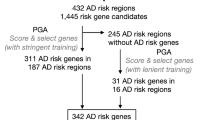
After initially collecting 335 AD-associated genes [Additional file 2], we then classified these genes into four categories, based on the strength of supporting evidence (the number of positive evidence of family-based studies and case-control studies). These were labeled C1-AD: probable pathogenic genes. C2-AD: high confidence genes. C3-AD: related genes, and C4-AD: possibly associated genes. Three hundred thirty-five AD-non associated genes as C5 group [Additional file 3]. Our goal is, within the scope of whole genome analysis, to find a stable relationship between a pair of genes, so as to find the AD candidate genes closely linked to genes known to be associated to AD.
According to the pathogenic mechanism of AD, we recruited the brain-specific gene network data from GIANT. Using these five gene groups, along with their evidence classification, and integrating brain-specific gene network data, we trained an evidence-weighted, network-based classifier, using an SVM approach. We randomly subdivided the total genes into two parts (10-fold cross-validation), which were used as training dataset and testing dataset, respectively. The classifier first identified network patterns (The relationship between any gene and 670 genes (335 AD associated genes +335 AD non-associated genes) as the features of SVM model). Then, we used the testing dataset to test the accuracy of the classifier, and divided them into initial two categories (AD related and AD non-related). We found that the average correct rate was 80.59% and the highest correct rate reached 84.56% with the ROC curve (receiver operating characteristic curve) as shown in Fig. 2. The ROC curve was constructed by “ROCR” package in R with a threshold of 0.561. Finally, we applied this classifier integrating the classification of known AD-associated genes to identify new AD candidate genes which interact closely with the known AD associated genes in the brain-specific network and divided those candidate genes into different groups by comparing the probability of each group and choosing the largest one. This method resulted in a comprehensive, genome-wide, ranked list of AD candidate genes.
To screen a more credible candidate gene list, we first annotated all AD associated genes (known AD-associated genes and AD predicted candidates) using the Gene Ontology resource (GO: http://www.geneontology.org/). We then performed a GO functional enrichment analysis of 335 known AD-associated genes and selected the top 10 GO items (Table 1) with P-values (adjusted by false discovery rate [19]) below a max value of 6.87e-11. Finally, we chose the AD predicted genes annotated on those GO items to further obtain high-confidence candidate genes. After this filter, we arrived at a total number of 832 AD predicted genes [Additional file 4].
The assessment of the full AD genes spectrum
In previous research, one prediction method has been based upon similarities in sequence-based features in disease genes [9,10,11]. Herein, a set of features was chosen from the genes, in order to assess our predictions.
The feature set (described in Table 2) reflects the structure and content of each gene examined. Table 3 lists the differences among the features present among the different sets of genes. Using the Mann-Whitney U test, we discovered highly significant differences in gene length, exon count, transcript count, 3’UTR length and 5’UTR length between the gene sequences of the AD-associated gene set and the control set of genes. Besides, there were also highly significant differences in transmembrane domain, signal domain and paralog calculated using the chi squared test. We reached the same conclusion when comparing the AD candidate dataset and control dataset.
Meanwhile, there was no statistically significant difference between the sequence patterns of AD-associated genes and AD candidate genes (as described in Table 4). The size (in bp) of the genes in both the AD-associated dataset and the AD candidate dataset is significantly larger than among controls, and this is similar to previous findings on AD association [9], which also report generally that genes associated with disease tend to be larger than those involved in normal phenotypes. In much the same way as larger total gene length is associated with AD, the length of 3’ UTR and 5’UTR, transcript count and the number of exons per gene, is in the AD-associated dataset and AD candidate dataset all larger. Genes in the known AD-associated dataset and the AD candidate dataset had a median count of 10 exons and 8 transcripts, while genes in the control dataset had a median count of 5 exons and 3 transcripts. We also found that there were significant differences in the length of 3’ UTR and 5’UTR in both AD-associated genes and AD candidate genes, which both had a larger median length of 3’UTR and 5’UTR, while genes in the control set had a smaller median length (described in Table 3).
Furtherly, we added genes annotated to non-mental-health diseases for comparison [14]. Differences calculated as p-value by using the Mann-Whitney U test or chi squared test between any two sets of four gene sets are shown in Table 4. On the basis of above results, we guessed that the differences between the sequence patterns of AD-associated genes and non-mental-health genes (as Group 1) were greater than that of AD-associated genes and AD candidate genes (as Group 2), but less than that of AD-associated genes and control genes (as Group 3). That is to say, our guess was that the p-values of Group 1 were smaller than that of Group 2, but larger than that of Group 3. Finally, most of the results were up to our expection, including length of gene and 5’ UTR, transcript count, transmembrane domain, signal domain and paralog.
Graphs presenting the distributions of each feature in the four datasets are shown in Fig. 3. It’s clear that the peaks of feature values of the known AD-associated dataset and AD candidate sets shift rightward, when compared to those of the control set. To our knowledge, the number of exons is also correlated to total gene length. However, the differences in 5’ UTR and 3’ UTR length have not been explained in terms of correlations to other feature differences.
Since the genes of the known AD-associated dataset were selected from the literature published before 2015, we also checked the predicted AD candidate gene set by scanning papers published post-2014 to verify the accuracy of our prediction (described in Table 5). As a result, we found that the AD candidate genes which were also reported in the post-2014 research could be roughly divided into several types. First, our gene candidates were identified as being associated with AD genes [20,21,22]. For example, DAB1, a novel candidate liability/protective gene, was identified by functional enrichment analysis of 3 AD Genome-Wide Association Studies (GWAS). Second, the genes were simply associated with risk of AD [23,24,25]. ANXA1 and CDC25C were identified as potentially contributing to AD susceptibility, by applying ICSNPathway analysis to the AD GWAS meta-analysis data. Third, abnormal changes in gene modification level or expression level were shown to exist in AD cases, when compared to the controls [23, 25]. For instance, DNA methylation levels within the CRTC1 gene were decreased in human hippocampus tissue affected by AD, suggesting that CRTC1 methylation plays an important role in AD pathophysiology.
Discussion
In this study, we have elucidated the whole-genome spectrum of AD, using a machine learning approach. We have classified the collected AD-associated genes by potential pathogenesis and taken advantage of brain-specific function networking to obtain correlations within the activity of any given pair of genes. Through this method, we expect for the candidate gene catalogue to provide a more comprehensive annotation of AD for researchers. Furthermore, this method could be applied to other brain disease pathogenic genes prediction, such as Parkinson’s disease, schizophrenia and so on.
By comparing the AD gene dataset with the control gene dataset in the sequence-based features (Tables 3 and 4), we found the median length of AD genes was much longer than controls. We speculate that the longer the gene is, the more mutations it will accumulate and the greater the pathogenicity it will be. Therefore, we suggest the research of related disease genes could be from the perspective of gene mutation load. What’s more, the proportion of genes with paralogs in the AD dataset is greater than in controls. It has been found human monogenic disease genes have frequently functionally redundant paralogs [26], but only one of the paralogous gene is associated with disease [27]. AD as a typical complex disease driven by multiple factors, the role of paralogs of pathogenic genes is worth further investigation.
The human brain has enormously complex cellular diversity, different parts of the neuron with different gene expression are specialized for different functions [28]. And AD is not caused by the role of single gene, so we need from the global point of view to study its development mechanism. The AD gene dataset obtained in our study can be used to explore the differences in gene expression and rare mutations distribution between AD patients and normal controls in different brain region, and clinically analyze the overexpression in AD brain neurons by single cell sequencing.
According to the GO functional enrichment analysis, we found AD-associated genes may play a role in the following GO items: protein binding, extracellular space, drug response, lipoprotein metabolic process, cholesterol metabolic process, cell surface, enzyme binding, beta-amyloid binding. To our best knowledge, most of them were concerned by different researchers, but there were no reports on drug response and enzyme binding, which may be a direction of our future study.
Conclusions
In this study, we have elucidated the whole-genome spectrum of AD, using a machine learning approach. Through this method, we expect for the candidate gene catalogue to provide a more comprehensive annotation of AD for researchers.
References
Talwar P, Sinha J, Grover S, Rawat C, Kushwaha S, Agarwal R, Taneja V, Kukreti R. Dissecting complex and multifactorial nature of Alzheimer's disease pathogenesis: a clinical, genomic, and systems biology perspective. Mol Neurobiol. 2016;53(7):4833–64.
Ulamek-Koziol M, Pluta R, Januszewski S, Kocki J, Bogucka-Kocka A, Czuczwar SJ. Expression of Alzheimer's disease risk genes in ischemic brain degeneration. Pharmacological reports : PR. 2016;68(6):1345–9.
Szigeti K. New genome-wide methods for elucidation of candidate copy number variations (CNVs) contributing to Alzheimer's disease heritability. Methods Mol Biol. 2016;1303:315–26.
Schadt EE, Lamb J, Yang X, Zhu J, Edwards S, Guhathakurta D, Sieberts SK, Monks S, Reitman M, Zhang C, et al. An integrative genomics approach to infer causal associations between gene expression and disease. Nat Genet. 2005;37(7):710–7.
Oti M, Snel B, Huynen MA, Brunner HG. Predicting disease genes using protein-protein interactions. J Med Genet. 2006;43(8):691–8.
Tang X, Hu X, Yang X, Fan Y, Li Y, Hu W, Liao Y, Zheng MC, Peng W, Gao L. Predicting diabetes mellitus genes via protein-protein interaction and protein subcellular localization information. BMC Genomics. 2016;17(Suppl 4):433.
Karni S, Soreq H, Sharan R. A network-based method for predicting disease-causing genes. Journal of computational biology : a journal of computational molecular cell biology. 2009;16(2):181–9.
Zhang Q, He M, Wang J, Liu S, Cheng H, Cheng Y. Predicting of disease genes for gestational diabetes mellitus based on network and functional consistency. Eur J Obstet Gynecol Reprod Biol. 2015;186:91–6.
Adie EA, Adams RR, Evans KL, Porteous DJ, Pickard BS. Speeding disease gene discovery by sequence based candidate prioritization. BMC bioinformatics. 2005;6:55.
Adie EA, Adams RR, Evans KL, Porteous DJ, Pickard BS. SUSPECTS: enabling fast and effective prioritization of positional candidates. Bioinformatics. 2006;22(6):773–4.
Lopez-Bigas N, Ouzounis CA. Genome-wide identification of genes likely to be involved in human genetic disease. Nucleic Acids Res. 2004;32(10):3108–14.
Zhang X, Acencio ML, Lemke N. Predicting essential genes and proteins based on machine learning and network topological features: a comprehensive review. Front Physiol. 2016;7:75.
Li M, Zhang J, Liu Q, Wang J, Wu FX. Prediction of disease-related genes based on weighted tissue-specific networks by using DNA methylation. BMC Med Genet. 2014;7(Suppl 2):S4.
Krishnan A, Zhang R, Yao V, Theesfeld CL, Wong AK, Tadych A, Volfovsky N, Packer A, Lash A, Troyanskaya OG. Genome-wide prediction and functional characterization of the genetic basis of autism spectrum disorder. Nat Neurosci. 2016;19(11):1454–62.
Stempler S, Yizhak K, Ruppin E. Integrating transcriptomics with metabolic modeling predicts biomarkers and drug targets for Alzheimer's disease. PLoS One. 2014;9(8):e105383.
Gomez Ravetti M, Moscato P. Identification of a 5-protein biomarker molecular signature for predicting Alzheimer's disease. PLoS One. 2008;3(9):e3111.
Ochagavia ME, Miranda J, Nazabal M, Martin A, Novoa LI, Bringas R, Fernandez DECJ, Camacho H. A methodology based on molecular interactions and pathways to find candidate genes associated to diseases: its application to schizophrenia and Alzheimer's disease. J Bioinforma Comput Biol. 2011;9(4):541–57.
Greene CS, Krishnan A, Wong AK, Ricciotti E, Zelaya RA, Himmelstein DS, Zhang R, Hartmann BM, Zaslavsky E, Sealfon SC, et al. Understanding multicellular function and disease with human tissue-specific networks. Nat Genet. 2015;47(6):569–76.
Chen JJ, Roberson PK, Schell MJ. The false discovery rate: a key concept in large-scale genetic studies. Cancer control : journal of the Moffitt Cancer Center. 2010;17(1):58–62.
Gao H, Tao Y, He Q, Song F, Saffen D. Functional enrichment analysis of three Alzheimer's disease genome-wide association studies identities DAB1 as a novel candidate liability/protective gene. Biochem Biophys Res Commun. 2015;463(4):490–5.
Shang Z, Lv H, Zhang M, Duan L, Wang S, Li J, Liu G, Ruijie Z, Jiang Y. Genome-wide haplotype association study identify TNFRSF1A, CASP7, LRP1B, CDH1 and TG genes associated with Alzheimer's disease in Caribbean Hispanic individuals. Oncotarget. 2015;6(40):42504–14.
Chen JA, Wang Q, Davis-Turak J, Li Y, Karydas AM, Hsu SC, Sears RL, Chatzopoulou D, Huang AY, Wojta KJ, et al. A multiancestral genome-wide exome array study of Alzheimer disease, frontotemporal dementia, and progressive supranuclear palsy. JAMA neurology. 2015;72(4):414–22.
Lee YH, Song GG. Genome-wide pathway analysis of a genome-wide association study on Alzheimer's disease. Neurol Sci. 2015;36(1):53–9.
Lin Q, Cao Y, Gao J. Decreased expression of the APOA1-APOC3-APOA4 gene cluster is associated with risk of Alzheimer's disease. Drug design, development and therapy. 2015;9:5421–31.
Chaudhry M, Wang X, Bamne MN, Hasnain S, Demirci FY, Lopez OL, Kamboh MI. Genetic variation in imprinted genes is associated with risk of late-onset Alzheimer's disease. Journal of Alzheimer's disease : JAD. 2015;44(3):989–94.
Chen W-H, Zhao X-M, van Noort V, Bork P. Human monogenic disease genes have frequently functionally redundant paralogs. PLoS Comput Biol. 2013;9(5):e1003073.
Adebali O, Reznik AO, Ory DS, Zhulin IB. Establishing the precise evolutionary history of a gene improves prediction of disease-causing missense mutations. Genetics in Medicine. 2016;18(10):1029.
Lake BB, Ai R, Kaeser GE, Salathia NS, Yung YC, Liu R, Wildberg A, Gao D, Fung H-L, Chen S. Neuronal subtypes and diversity revealed by single-nucleus RNA sequencing of the human brain. Science. 2016;352(6293):1586–90.
Aken BL, Ayling S, Barrell D, Clarke L, Curwen V, Fairley S, Fernandez Banet J, Billis K, Garcia Giron C, Hourlier T et al. The Ensembl gene annotation system. Database : the journal of biological databases and curation. 2016;2016:baw093.
UniProt C. UniProt: a hub for protein information. Nucleic Acids Res. 2015;43(Database issue):D204–12.
Xiao Q, Liu Z-J, Tao S, Sun Y-M, Jiang D, Li H-L, Chen H, Liu X, Lapin B, Wang C-H. Risk prediction for sporadic Alzheimer's disease using genetic risk score in the Han Chinese population. Oncotarget. 2015;6(35):36955.
Malishkevich A, Marshall GA, Schultz AP, Sperling RA, Aharon-Peretz J, Gozes I. Blood-borne activity-dependent neuroprotective protein (ADNP) is correlated with premorbid intelligence, clinical stage, and Alzheimer’s disease biomarkers. J Alzheimers Dis. 2016;50(1):249-60.
Zheng X, Demirci F, Barmada M, Richardson G, Lopez O, Sweet R, Kamboh M, Feingold E. Genome-wide copy-number variation study of psychosis in Alzheimer’s disease. Transl Psychiatry. 2015;5(6):e574.
Marchesi VT. Gain-of-function somatic mutations contribute to inflammation and blood vessel damage that lead to Alzheimer dementia: a hypothesis. FASEB J. 2016;30(2):503-6.
Acknowledgements
We are grateful to the technical staff at BGI-Shenzhen for their assistance and technical support. We would like to thank Shenzhen Key Laboratory of Neurogenomics (BGI-Shenzhen) and Alzheimer’s disease project. We thank for the support of Shenzhen Municipal of Government of China (NO: CXB201108250094A and JSGG2015330171719763) and National Natural Science Foundation of China (NO: Z151100003915117).
Funding
This work is supported by the Shenzhen Municipal Government of China (No: CXB201108250094A and JSGG2015330171719763) and National Natural Science Foundation of China (NO: Z151100003915117). The funders had no role in the design of the study, data collection, analysis, and interpretation, or in writing the manuscript.
Availability of data and materials
Authors can confirm that all relevant data are included in the article and/or its supplementary information files.
Author information
Authors and Affiliations
Contributions
XH contributed to perform data analysis, and was a major contributor in writing the manuscript. HL contributed to the design of the study and wrote the manuscript. XL contributed to perform data analysis and was a contributor in writing the manuscript. LG and LJ contributed to data analysis and helped with the manuscript. LT contributed to modify the article. HY and JW contributed to provide computational resources support for the research and give final approval of the manuscript. JZ contributed to the design of the study, and helped with the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Additional files
Additional file 1: Figure S1.
The correct rates of different non-associated gene sets in SVM training. We randomly selected the dataset of 335 non-associated genes (n = 100) for SVM training. (TIFF 12473 kb)
Additional file 2:
335 AD-associated genes. The datasets collected from public Alzheimer’s disease databases (AlzGene) and the publications treating upon AD. (XLSX 14 kb)
Additional file 3:
The classification of AD-associated genes and AD-non associated genes. C1-AD: probable pathogenic genes; C2-AD: high confidence genes; C3-AD: related genes; C4-AD: possibly associated genes; C5-AD: AD-non associated genes. (XLSX 19 kb)
Additional file 4:
832 AD predicted genes. A total number of AD predicted genes across the whole genome in our study. (XLSX 68 kb)
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Huang, X., Liu, H., Li, X. et al. Revealing Alzheimer’s disease genes spectrum in the whole-genome by machine learning. BMC Neurol 18, 5 (2018). https://doi.org/10.1186/s12883-017-1010-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12883-017-1010-3