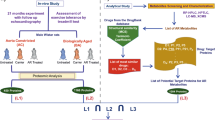
Abstract
Mucopolysaccharidoses are caused by a deficiency of enzymes involved in the degradation of glycosaminoglycans. Heart diseases are a significant cause of morbidity and mortality in MPS patients, even in conditions in which enzyme replacement therapy is available. In this sense, cardiovascular manifestations, such as heart hypertrophy, cardiac function reduction, increased left ventricular chamber, and aortic dilatation, are among the most frequent. However, the downstream events which influence the heart dilatation process are unclear. Here, we employed systems biology tools together with transcriptomic data to investigate new elements that may be involved in aortic dilatation in Mucopolysaccharidoses syndrome. We identified candidate genes involved in biological processes related to inflammatory responses, deposition of collagen, and lipid accumulation in the cardiovascular system that may be involved in aortic dilatation in the Mucopolysaccharidoses I and VII. Furthermore, we investigated the molecular mechanisms of losartan treatment in Mucopolysaccharidoses I mice to underscore how this drug acts to prevent aortic dilation. Our data indicate that the association between the TGF-b signaling pathway, Fos, and Col1a1 proteins can play an essential role in aortic dilation's pathophysiology and its subsequent improvement by losartan treatment.




Similar content being viewed by others
References
Giugliani R (2012) Mucopolysacccharidoses: From understanding to treatment, a century of discoveries. Genet Mol Biol 35:924–931. https://doi.org/10.1590/S1415-47572012000600006
Stapleton M, Arunkumar N, Kubaski F et al (2018) Clinical presentation and diagnosis of mucopolysaccharidoses. Mol Genet Metab 125:4–17. https://doi.org/10.1016/j.ymgme.2018.01.003
Muenzer J (2004) The mucopolysaccharidoses: a heterogeneous group of disorders with variable pediatric presentations. J Pediatr 144:27–34. https://doi.org/10.1016/j.jpeds.2004.01.052
Stapleton M, Hoshina H, Sawamoto K et al (2019) Critical review of current MPS guidelines and management. Mol Genet Metab 126:238–245. https://doi.org/10.1016/j.ymgme.2018.07.001
Matte U, Yogalingam G, Brooks D et al (2003) Identification and characterization of 13 new mutations in mucopolysaccharidosis type I patients. Mol Genet Metab 78:37–43. https://doi.org/10.1016/S1096-7192(02)00200-7
Yogalingam G, Guo X-H, Muller VJ et al (2004) Identification and molecular characterization of alpha-L-iduronidase mutations present in mucopolysaccharidosis type I patients undergoing enzyme replacement therapy. Hum Mutat 24:199–207. https://doi.org/10.1002/humu.20081
Pollard LM, Jones JR, Wood TC (2013) Molecular characterization of 355 mucopolysaccharidosis patients reveals 104 novel mutations. J Inherit Metab Dis 36:179–187. https://doi.org/10.1007/s10545-012-9533-7
Lin HY, Lin SP, Chuang CK et al (2005) Mucopolysaccharidosis I under enzyme replacement therapy with laronidase—a mortality case with autopsy report. J Inherit Metab Dis 28:1146–1148. https://doi.org/10.1007/s10545-005-0211-x
Martins AM, Dualibi AP, Norato D et al (2009) Guidelines for the management of mucopolysaccharidosis type I. J Pediatr. https://doi.org/10.1016/j.jpeds.2009.07.005
Imundo L, LeDuc CA, Guha S et al (2011) A complete deficiency of Hyaluronoglucosaminidase 1 (HYAL1) presenting as familial juvenile idiopathic arthritis. J Inherit Metab Dis 34:1013–1022. https://doi.org/10.1007/s10545-011-9343-3
Poswar FdO, de Souza CFM, Giugliani R, Baldo G (2019) Aortic root dilatation in patients with mucopolysaccharidoses and the impact of enzyme replacement therapy. Heart Vessels 34:290–295. https://doi.org/10.1007/s00380-018-1242-1
Braunlin EA, Harmatz PR, Scarpa M et al (2011) Cardiac disease in patients with mucopolysaccharidosis: presentation, diagnosis and management. J Inherit Metab Dis 34:1183–1197. https://doi.org/10.1007/s10545-011-9359-8
Bolourchi M, Renella P, Wang RY (2016) Aortic root dilatation in mucopolysaccharidosis I–VII. Int J Mol Sci 17:3–9. https://doi.org/10.3390/ijms17122004
Bigg PW, Baldo G, Sleeper MM et al (2013) Pathogenesis of mitral valve disease in mucopolysaccharidosis VII dogs. Mol Genet Metab 110:319–328. https://doi.org/10.1016/j.ymgme.2013.06.013
Jurecka A, Tylki-Szymańska A (2015) Enzyme replacement therapy: lessons learned and emerging questions. Expert Opin Orphan Drugs 3:293–305. https://doi.org/10.1517/21678707.2015.1017469
Baldo G, Tavares AMV, Gonzalez E et al (2017) Progressive heart disease in mucopolysaccharidosis type I mice may be mediated by increased cathepsin B activity. Cardiovasc Pathol 27:45–50. https://doi.org/10.1016/j.carpath.2017.01.001
Lew WYW, Bayna E, Molle ED et al (2013) Recurrent exposure to subclinical lipopolysaccharide increases mortality and induces cardiac fibrosis in mice. PLoS ONE 8:1–11. https://doi.org/10.1371/journal.pone.0061057
Simonaro CM, D’Angelo M, He X et al (2008) Mechanism of glycosaminoglycan-mediated bone and joint disease: implications for the mucopolysaccharidoses and other connective tissue diseases. Am J Pathol 172:112–122. https://doi.org/10.2353/ajpath.2008.070564
Ausseil J, Desmaris N, Bigou S et al (2008) Early neurodegeneration progresses independently of microglial activation by heparan sulfate in the brain of mucopolysaccharidosis IIIB mice. PLoS ONE. https://doi.org/10.1371/journal.pone.0002296
Baldo G, Wu S, Howe RA et al (2011) Pathogenesis of aortic dilatation in mucopolysaccharidosis VII mice may involve complement activation. Mol Genet Metab 104:608–619. https://doi.org/10.1016/j.ymgme.2011.08.018
Ma X, Tittiger M, Knutsen RH et al (2008) Upregulation of elastase proteins results in aortic dilatation in mucopolysaccharidosis I mice. Mol Genet Metab 94:298–304. https://doi.org/10.1016/j.ymgme.2008.03.018
Gonzalez EA, Tavares AMV, Poletto E et al (2017) Losartan improves aortic dilatation and cardiovascular disease in mucopolysaccharidosis I. J Inherit Metab Dis 40:311–312. https://doi.org/10.1007/s10545-017-0014-x
Sean D, Meltzer PS (2007) GEOquery: a bridge between the Gene Expression Omnibus (GEO) and BioConductor. Bioinformatics 23:1846–1847. https://doi.org/10.1093/bioinformatics/btm254
Du P, Kibbe WA, Lin SM (2008) lumi: a pipeline for processing Illumina microarray. Bioinformatics 24:1547–1548. https://doi.org/10.1093/bioinformatics/btn224
Dunning MJ, Smith ML, Ritchie ME, Tavaré S (2007) Beadarray: R classes and methods for Illumina bead-based data. Bioinformatics 23:2183–2184. https://doi.org/10.1093/bioinformatics/btm311
Kauffmann A, Gentleman R, Huber W (2009) arrayQualityMetrics—a bioconductor package for quality assessment of microarray data. Bioinformatics 25:415–416. https://doi.org/10.1093/bioinformatics/btn647
Smyth GK (2005) limma: Linear models for microarray data. In: Gentleman R, Carey VJ, Huber W, Irizarry RA, Dudoit S (eds) Bioinformatics and computational biology solutions using R and Bioconductor. Springer, New York, pp 397–420
Shannon P, Markiel A, Ozier O et al (2003) Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. https://doi.org/10.1101/gr.1239303.metabolite
Scardoni G, Petterlini M, Laudanna C (2009) Analyzing biological network parameters with CentiScaPe. Bioinformatics 25:2857–2859. https://doi.org/10.1093/bioinformatics/btp517
Pavlopoulos GA, Secrier M, Moschopoulos CN et al (2011) Using graph theory to analyze biological networks. BioData Min 4:1–27. https://doi.org/10.1186/1756-0381-4-10
Yu H, Kim PM, Sprecher E et al (2007) The importance of bottlenecks in protein networks: correlation with gene essentiality and expression dynamics. PLoS Comput Biol 3:713–720. https://doi.org/10.1371/journal.pcbi.0030059
Menche J, Guney E, Sharma A et al (2017) Integrating personalized gene expression profiles into predictive disease-associated gene pools. NPJ Syst Biol Appl. https://doi.org/10.1038/s41540-017-0009-0
Kitsak M, Havlin S, Paul G et al (2007) Betweenness centrality of fractal and nonfractal scale-free model networks and tests on real networks. Phys Rev E 75:5. https://doi.org/10.1103/PhysRevE.75.056115
Bader GD, Hogue CWV (2003) An automated method for finding molecular complexes in large protein interaction networks. BMC Bioinformatics 4:1–27. https://doi.org/10.1186/1471-2105-4-2
Maere S, Heymans K, Kuiper M (2005) BiNGO: a cytoscape plugin to assess overrepresentation of gene ontology categories in biological networks. Bioinformatics 21:3448–3449. https://doi.org/10.1093/bioinformatics/bti551
Benjamini Y, Hochberg Y (1995) Controlling the false discovery rate: a practical approach to multiple testing. J Royal Stat Soc B 57:289–300. https://www.jstor.org/stable/2346101
Menche J, Sharma A, Kitsak M et al (2015) Uncovering disease-disease relationships through the incomplete interactome. Science (80-) 347:841. https://doi.org/10.1126/science.1257601
urali TM, Gil DP, Law JN (2017) The PathLinker app: Connect the dots in protein interaction networks. F1000Research 6:58
Ritz A, Poirel CL, Tegge AN et al (2016) Pathways on demand: automated reconstruction of human signaling networks. NPJ Syst Biol Appl. https://doi.org/10.1038/npjsba.2016.2
Barabási AL, Oltvai ZN (2004) Network biology: Understanding the cell’s functional organization. Nat Rev Genet 5:101–113. https://doi.org/10.1038/nrg1272
Watts DJ, Strogatz SH (1998) Strogatz - small world network Nature. Nature 393:440–442
Schadt EE (2009) Molecular networks as sensors and drivers of common human diseases. Nature 461:218–223. https://doi.org/10.1038/nature08454
Barabási AL, Gulbahce N, Loscalzo J (2011) Network medicine: a network-based approach to human disease. Nat Rev Genet 12:56–68. https://doi.org/10.1038/nrg2918
Caldera M, Buphamalai P, Müller F, Menche J (2017) Interactome-based approaches to human disease. Curr Opin Syst Biol 3:88–94. https://doi.org/10.1016/j.coisb.2017.04.015
Woidy M, Muntau AC, Gersting SW (2018) Inborn errors of metabolism and the human interactome: a systems medicine approach. J Inherit Metab Dis 41:285–296. https://doi.org/10.1007/s10545-018-0140-0
Il GK, Cusick ME, Valle D et al (2007) The human disease network. Proc Natl Acad Sci U S A 104:8685–8690. https://doi.org/10.1073/pnas.0701361104
Feldman I, Rzhetsky A, Vitkup D (2008) Network properties of genes harboring inherited disease mutations. Proc Natl Acad Sci U S A 105:4323–4328. https://doi.org/10.1073/pnas.0701722105
Metcalf JA, Linders B, Wu S et al (2010) Upregulation of elastase activity in aorta in mucopolysaccharidosis I and VII dogs may be due to increased cytokine expression. Mol Genet Metab 99:396–407. https://doi.org/10.1016/j.ymgme.2009.12.003
Khalid O, Vera MU, Gordts PL et al (2016) Immune-mediated inflammation may contribute to the pathogenesis of cardiovascular disease in mucopolysaccharidosis Type I. PLoS ONE 11:e0150850. https://doi.org/10.1371/journal.pone.0150850
Sleeper MM, Fornasari B, Ellinwood NM et al (2004) Gene therapy ameliorates cardiovascular disease in dogs with mucopolysaccharidosis VII. Circulation 110:815–820. https://doi.org/10.1161/01.CIR.0000138747.82487.4B
Newman MEJ (2006) Modularity and community structure in networks. Proc Natl Acad Sci 103:8577–8582. https://doi.org/10.1073/pnas.0601602103
Alfonso Pecchio AR, Cardozo Gizzi AM, Renner ML et al (2011) c-Fos activates and physically interacts with specific enzymes of the pathway of synthesis of polyphosphoinositides. Mol Biol Cell 22:4716–4725. https://doi.org/10.1091/mbc.E11-03-0259
Maruyama K, Sano GI, Ray N et al (2007) C-fos-deficient mice are susceptible to Salmonella enterica serovar typhimurium infection. Infect Immun 75:1520–1523. https://doi.org/10.1128/IAI.01316-06
Alfano D, Franco P, Vocca I et al (2005) The urokinase plasminogen activator and its receptor role in cell growth and apoptosis. Thromb Haemost 93:205–211. https://doi.org/10.1160/TH04
Takaya N, Katoh Y, Iwabuchi K et al (2005) Platelets activated by collagen through the immunoreceptor tyrosine-based activation motif in the Fc receptor γ-chain play a pivotal role in the development of myocardial ischemia-reperfusion injury. J Mol Cell Cardiol 39:856–864. https://doi.org/10.1016/j.yjmcc.2005.07.006
Srikakulapu P, Hu D, Yin C et al (2016) Artery tertiary lymphoid organs control multilayered territorialized atherosclerosis B-cell responses in Aged ApoE-/- mice. Arterioscler Thromb Vasc Biol 36:1174–1185. https://doi.org/10.1161/ATVBAHA.115.306983
Araujo AC, Marques S, Belo JA (2014) Targeted inactivation of cerberus like-2 leads to left ventricular cardiac hyperplasia and systolic dysfunction in the mouse. PLoS ONE. https://doi.org/10.1371/journal.pone.0102716
Giraud MN, Flück M, Zuppinger C, Suter TM (2005) Expressional reprogramming of survival pathways in rat cardiocytes by neuregulin-1β. J Appl Physiol 99:313–322. https://doi.org/10.1152/japplphysiol.00609.2004
Hajishengallis G, Lambris JD (2010) Crosstalk pathways between toll-like receptors and the complement system. Trends Immunol 31:154–163. https://doi.org/10.1016/j.it.2010.01.002
Prentice RL, Zhao S, Johnson M et al (2013) Proteomic risk markers for coronary heart disease and stroke: validation and mediation of randomized trial hormone therapy effects on these diseases. Genome Med. https://doi.org/10.1186/gm517
Chiu HC, Kovacs A, Ford DA et al (2001) A novel mouse model of lipotoxic cardiomyopathy. J Clin Invest 107:813–822. https://doi.org/10.1172/JCI10947
Boyle EA, Li YI, Pritchard JK (2017) An expanded view of complex traits: from polygenic to omnigenic. Cell 169:1177–1186. https://doi.org/10.1016/j.cell.2017.05.038
Goldberg IJ, Trent CM, Schulze PC (2012) Lipid metabolism and toxicity in the heart. Cell Metab 15:805–812. https://doi.org/10.1016/j.cmet.2012.04.006
Yang L, Yang Y, Si D et al (2017) High expression of long chain acyl-coenzyme a synthetase 1 in peripheral blood may be a molecular marker for assessing the risk of acute myocardial infarction. Exp Ther Med 14:4065–4072. https://doi.org/10.3892/etm.2017.5091
Cleutjens JPM, Creemers EEJM (2002) Integration of concepts: cardiac extracellular matrix remodeling after myocardial infarction. J Card Fail 8:344–348. https://doi.org/10.1054/jcaf.2002.129261
Yim J, Cho H, Rabkin SW (2018) Gene expression and gene associations during the development of heart failure with preserved ejection fraction in the Dahl salt sensitive model of hypertension. Clin Exp Hypertens 40:155–166. https://doi.org/10.1080/10641963.2017.1346113
Tian HP, Sun YH, He L et al (2018) Single-stranded DNA-binding protein 1 abrogates cardiac fibroblast proliferation and collagen expression induced by angiotensin II. Int Heart J 59:1398–1408. https://doi.org/10.1536/ihj.17-650
Siddesha JM, Valente AJ, Sakamuri SSVP et al (2013) Angiotensin II stimulates cardiac fibroblast migration via the differential regulation of matrixins and RECK. J Mol Cell Cardiol 65:9–18. https://doi.org/10.1016/j.yjmcc.2013.09.015
Gallini R, Lindblom P, Bondjers C et al (2016) PDGF-A and PDGF-B induces cardiac fibrosis in transgenic mice. Exp Cell Res 349:282–290. https://doi.org/10.1016/j.yexcr.2016.10.022
Chang S, McKinsey TA, Zhang CL et al (2004) Histone deacetylases 5 and 9 govern responsiveness of the heart to a subset of stress signals and play redundant roles in heart development. Mol Cell Biol 24:8467–8476. https://doi.org/10.1128/mcb.24.19.8467-8476.2004
Monovich L, Koch KA, Burgis R et al (2009) Suppression of HDAC nuclear export and cardiomyocyte hypertrophy by novel irreversible inhibitors of CRM1. Biochim Biophys Acta Gene Regul Mech 1789:422–431. https://doi.org/10.1016/j.bbagrm.2009.04.001
Xu WP, Yao TQ, Jiang YB et al (2015) Effect of the angiotensin ii receptor blocker valsartan on cardiac hypertrophy and myocardial histone deacetylase expression in rats with aortic constriction. Exp Ther Med 9:2225–2228. https://doi.org/10.3892/etm.2015.2374
Osborn MJ, Webber BR, McElmurry RT et al (2017) Angiotensin receptor blockade mediated amelioration of mucopolysaccharidosis type I cardiac and craniofacial pathology. J Inherit Metab Dis 40:281–289. https://doi.org/10.1007/s10545-016-9988-z
Chen Y, Lasaitiene D, Gabrielsson BG et al (2004) Neonatal losartan treatment suppresses renal expression of molecules involved in cell–cell and cell–matrix interactions. J Am Soc Nephrol 15:1232–1243. https://doi.org/10.1097/01.ASN.0000123690.75029.3F
Tsai CT, Lai LP, Hwang JJ et al (2008) Renin-angiotensin system component expression in the HL-1 atrial cell line and in a pig model of atrial fibrillation. J Hypertens 26:570–582. https://doi.org/10.1097/HJH.0b013e3282f34a4a
Crews EC, Rowland NE (2005) Role of angiotensin in body fluid homeostasis of mice: effect of losartan on water and NaCl intakes. Am J Physiol Regul Integr Comp Physiol 288:638–644. https://doi.org/10.1152/ajpregu.00525.2004
Zacchigna L, Vecchione C, Notte A et al (2006) Emilin1 links TGF-β maturation to blood pressure homeostasis. Cell 124:929–942. https://doi.org/10.1016/j.cell.2005.12.035
Kang JS, Alliston T, Delston R, Derynck R (2005) Repression of Runx2 function by TGF-β through recruitment of class II histone deacetylases by Smad3. EMBO J 24:2543–2555. https://doi.org/10.1038/sj.emboj.7600729
Molina JR, Adjei AA (2006) The Ras/Raf/MAPK pathway. J Thorac Oncol 1:7–9. https://doi.org/10.1016/s1556-0864(15)31506-9
Derynck R, Budi EH (2019) Specificity, versatility, and control of TGF-b family signaling. Sci Signal. https://doi.org/10.1126/scisignal.aav5183
Massagué J, Seoane J, Wotton D (2005) Smad transcription factors. Genes Dev 19:2783–2810. https://doi.org/10.1101/gad.1350705
Wang J, Xu N, Feng X et al (2005) Targeted disruption of Smad4 in cardiomyocytes results in cardiac hypertrophy and heart failure. Circ Res 97:821–828. https://doi.org/10.1161/01.RES.0000185833.42544.06
Xu J, Gruber PJ, Chien KR (2019) SMAD4 is essential for human cardiac mesodermal precursor cell formation. Stem Cells 37:216–225. https://doi.org/10.1002/stem.2943
Funding
T.C. was supported by Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq). B.C.F. was also supported by CNPq (151680/2019-1).
Author information
Authors and Affiliations
Contributions
TC, BCF, and UM conceived and designed the study, TC and BCF formal analysis, data curation, and wrote the manuscript. EG and GB helped in analyzing data. UM, and BCF revised the manuscript. All authors read and approved the final version of the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Corrêa, T., Feltes, B.C., Gonzalez, E.A. et al. Network Analysis Reveals Proteins Associated with Aortic Dilatation in Mucopolysaccharidoses. Interdiscip Sci Comput Life Sci 13, 34–43 (2021). https://doi.org/10.1007/s12539-020-00406-3
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12539-020-00406-3