Abstract
Metabarcoding is a rapidly developing tool in marine zooplankton ecology, although most zooplankton surveys continue to rely on visual identification for monitoring purposes. We attempted to resolve some of the biases associated with metabarcoding by sequencing a 313-b.p. fragment of the COI gene in 34 “mock” samples from the North Sea which were pre-sorted to species level, with biomass and abundance estimates obtained for each species and taxonomic group. The samples were preserved either in 97% ethanol or dehydrated for 24 h in a drying oven at 65 °C (the routine way of preserving samples for dry weight measurements). The visual identification yielded a total of 59 unique holoplanktonic and 16 meroplanktonic species/taxa. Metabarcoding identified 86 holoplanktonic and 124 meroplanktonic species/taxa, which included all but 3 of the species identified visually as well as numerous species of hard-to-identify crustaceans, hydrozoan jellyfish, and larvae of benthic animals. On a sample-to-sample basis, typically 90–95% of visually registered species were recovered, but the number of false positives was also high. We demonstrate robust correlations of relative sequence abundances to relative biomass for most taxonomic groups and develop conversion factors for different taxa to account for sequencing biases. We then combine the adjusted sequencing data with a single bulk biomass measurement for the entire sample to produce a quantitative parameter akin to species biomass. When examined with multivariate statistics, this parameter, which we call BWSR (biomass-weighed sequence reads) showed very similar trends to species biomass and comparable patterns to species abundance, highlighting the potential of metabarcoding not only for biodiversity estimation and mapping of presence/absence of species but also for quantitative assessment of zooplankton communities.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Marine zooplankton play a crucial role in ocean ecosystems, linking primary producers to higher trophic levels and contributing to the biogeochemical cycling of nutrients. They are also indicators of the health of marine environments and can provide valuable insights into the impacts of climate change, pollution, and other anthropogenic activities on the ocean (Ferdous and Muktadir 2009; Ndah et al. 2022; Yang and Zhang 2020). However, many of the ecologically significant indicators, such as the presence or relative abundance of sensitive or invasive species, rely on accurate species-level data, which remains a bottleneck in many zooplankton studies, given the time and taxonomic expertise required to manually sort and identify zooplankton samples.
Recent advances in molecular techniques have allowed to study marine biological communities at an unprecedented volume and taxonomic resolution and are rapidly becoming more and more affordable. More specifically, metabarcoding, or the simultaneous sequencing of a DNA barcode region of interest in all organisms in a community sample, permits the analysis of 100 s to 1000 s of samples simultaneously (Gaither et al. 2022). Metabarcoding avoids a lot of the limitations of microscopic sorting and more modern visual-based identification methods, since it can distinguish organisms of any stage of development, cryptic species, or those species that lose their distinguishing features in preservatives. Metabarcoding can also be more cost-effective than traditional methods, particularly for large-scale monitoring studies, as it requires fewer trained personnel and can be to a large extent automated. However, there are still methodological challenges and limitations that must be properly addressed, including the choice of barcode, PCR biases, and incomplete reference libraries (Bucklin et al. 2016; Santoferrara 2019). Most importantly, despite a growing body of evidence that sequence numbers produced via metabarcoding are at least somewhat representative of the relative composition of the organisms in the environment, it remains a challenge to apply metabarcoding data for community analysis in a quantitative way (Bucklin et al. 2016). The most common quantitative parameter in plankton ecology, abundance of organisms, is expectedly often poorly correlated to sequence numbers due to the enormous range in organism size and, thus, their DNA content. This can even include organisms belonging to the same species—for example, a Calanus spp. individual will vary between 0.2 and 140 µg C, or 4 orders of magnitude, between an egg and its adult stage (i.e., Møller et al. 2016). Far better correlations have been observed for organism biomass, especially in certain groups such as copepods (Ershova et al. 2021; Hirai et al. 2017; Lamb et al. 2019; Matthews et al. 2021), although typically these relationships remain far from perfect for most taxa, presumably due to PCR bias as well as different DNA density between taxonomic groups of similar weight.
In this study, we apply a mock sample approach to quantify and address the biases of metabarcoding both for recovering biodiversity and estimating relative contribution of species. We capitalize on the well-documented relationship between organism biomass and sequence numbers (Ershova et al. 2021; Hirai et al. 2017; Krehenwinkel et al. 2017; Schenk et al. 2019) to introduce a direct framework to apply metabarcoding as a quantitative method in zooplankton monitoring studies. We sequenced a 313-base pair fragment (“Leray” fragment) of the COI barcoding gene, which has been shown to be successful in recovering biodiversity at the species level in a wide range of marine invertebrates, including zooplankton (Antich et al. 2019; Ershova et al. 2021; Wangensteen et al. 2018). We attempt to correct for some of the PCR bias and other factors that result in uneven sequencing of various organisms, with the hypothesis that under- or overrepresentation of specific taxa in sequencing data is consistent and predictable. We do this by developing species- or taxa- specific conversion factors that are applied across the entire dataset and bring the relationship between organism biomass and sequence reads closer to unity. We then combine this data with a single bulk biomass measurement for the entire sample to produce a quantitative molecular variable akin to species biomass, which can then be used to map species distributions and densities, analyze community structure, and estimate a variety of ecosystem indicators. Additionally, we tested two different methods of preservation of zooplankton DNA to evaluate the impact of preservation method on the obtained results. We propose that the protocol described in this work can be a valuable tool for future studies on marine zooplankton communities and to help better understand the functioning of marine ecosystems.
Materials and methods
Collection and sample preparation
Sampling and mock sample preparation were done during the IMR North Sea Ecosystem Cruise onboard the RV Johan Hjort during April 2022 (Fig. 1, Supplementary Material 1). Zooplankton was collected using a WP-II net (0.25-m2 mouth opening, 180-µm mesh size), which was hauled vertically from 5 m off the sea floor. At two of the stations (stations 418 and 423), samples were obtained using a Multinet MAMMOTH (Hydrobios) with a 180-µm mesh containing 9 nets, hauled obliquely from 5 m above the sea floor while the boat was moving at a speed of 1 km. Additionally, at almost every station, supplementary samples were obtained using the GULF VII net with a mesh size of 280 µm which was also hauled obliquely from 100-m depth.
A small subsample (typically, 1/8 of the total sample) was obtained from the WP-II nets (16 stations) or the deepest net of the MultiNet MAMMOTH (2 stations). The animals were immobilized using a few drops of carbonated sea water then sorted under a stereomicroscope, and 200–500 animals were selected to create a “mock” sample. The sample was also supplemented with larger and/or rarer animals from the GULF VII net from the corresponding station. Per this method, the diversity within the samples reflected the real environmental diversity at that location (i.e., only animals from a given station were used to create the sample), but their relative abundances were to a large degree artificial. The samples were created to maximize diversity and varying contribution of various taxonomic groups in different samples. Organisms were identified to species level when possible, although for many organisms identification was done at the genus level or higher, especially at larval/juvenile stages. For example, copepods belonging to the genera Pseudocalanus and Paracalanus were identified as Pseudo/Paracalanus spp., and Calanus finmarchicus and C. helgolandicus were grouped as Calanus spp. All calanoid copepod nauplii were identified as Calanoida. Meroplankton was typically grouped at the phylum or class level. The body length of each animal was measured using the ZoopBiom digitizing system (Roff and Hopcroft 1986), and its biomass was estimated using a length–weight regression relationship published for the species or a similar species (see Ershova et al. 2015 for details). While this indirect method of biomass estimation has an associated bias, especially for groups for which there is limited data on length–weight relationships, it allows to estimate species weight without the labor-intensive practice of weighing every group of organisms. Total sample biomass was estimated by summing all the individual dry weight values. Nitrile gloves were worn during sample preparation, and all dishes and sorting tools were rinsed with 10% bleach between samples to avoid contamination. At most locations, two samples were created for each station: one was preserved in 100% ethanol, and the other was rinsed with freshwater and placed on a pre-weighed aluminum tray, then dried at 65 °C for 24 h. These two samples per station were designed to be very similar in quantity and composition, but were not identical. In total, 33 samples were prepared from 18 stations (17 preserved via drying, and 16 in ethanol).
DNA analysis
In the lab, the dry samples were re-constituted with a few drops of MilliQ water and ethanol was drained and replaced with MilliQ water. The samples were homogenized in 2-ml tubes containing ceramic beads using a 2 × 150 Precellys machine. Three 100-µl replicates of the resulting homogenate were taken, and DNA was extracted from each replicate using the Qiagen Blood and Tissue Kit according the manufacturer’s protocol. PCR amplification was carried out using individually tagged Leray-XT primers for COI: forward primer mlCOIintF-XT 5'-GGWACWRGWTGRACWITITAYCCYCC-3' (Wangensteen et al. 2018) and reverse primer jgHCO2198 5'-TAIACYTCIGGRTGICCRAARAAYCA-3' (Geller et al. 2013). The reagents and PCR protocol are described in detail in Ershova et al. 2021. Two extraction blanks and two PCR blanks were sequenced with the samples as negative controls. The PCR products were visualized on a gel to ensure the absence of contamination, then purified using Minelute PCR purification columns (www.qiagen.com) and pooled into a single library. The NextFlex PCR-free library preparation kit (Perkin-Elmer) was used to prepare the Illumina library, which was then sequenced on an Illumina MiSeq using ½ of a V3 2 × 250-bp kit (Illumina).
Bioinformatics
The bioinformatics pipeline closely followed Ershova et al. 2021. Paired-end reads were aligned with illuminapairedend from OBITools v1.01.22 (Boyer et al. 2016), and reads with an alignment score < 40 were discarded. Primer sequences were removed, and reads were demultiplexed and assigned to individual samples using ngsfilter. Reads were selected for length (299–320 b.p.) using obigrep and dereplicated using obiuniq. The uchime_denovo algorithm (Edgar et al. 2011) implemented in vsearch v1.10.1 (Rognes et al. 2016) was then used to remove chimeric sequences. Singletons (sequences with abundance of 1 read) were removed, and step-by-step clustering was performed in SWARM 3.0.0 (Mahé et al. 2021) using a distance value of d = 13, which is the optimal value for this marker (Antich et al. 2021), to cluster individual sequences into molecular operational taxonomic units (MOTUs). Taxonomic assignment of the representative sequences was then performed using ecotag (Boyer et al. 2016) against DUFA-Leray v.2020–06-10, a custom reference database (publicly available from github.com/uit-metabarcoding/DUFA), which included sequences of the Leray fragment extracted from BOLD and Genbank, complemented with in-house generated sequences. This method uses a phylogenetic tree-based approach, and when a sequence fails to produce a perfect match, it identifies taxa which are at least as similar to the nearest hit as the sequence in question, and assigns it to its most recent common ancestor, usually resulting in a higher rank, such as genus, family, or order. In the cases when not enough related sequences were present in the database to perform this analysis, an artificial threshold was applied (95% similarity for genus, 90% for family, 85% for order, and 75% for class). The resulting dataset was curated for putative pseudogene sequences using LULU (Frøslev et al. 2017). The next refining steps consisted of removing MOTUs assigned to prokaryotes and non-planktonic organisms (e.g., mammals) and a second manual taxonomy check of all MOTUs using BOLD (Barcode of Life Database, www.boldsystems.org) using species-level barcode records. A species-level identification was assigned with a minimum of 97% similarity. Several MOTU’s produced more than one species-level match and were dealt with on a case-to-case basis, with credibility of reference sources and known species distributions playing the main role in the final assignment. After all the bioinformatics steps, the three extraction replicates were pooled for each sample.
Data analysis
All analyses were performed in R (R Development Core Team 2011). Pearson correlation coefficients (r) were calculated between relative biomass, abundance, and sequence reads of all taxa at the lowest common taxonomic resolution both for the entire dataset and for each sample individually. Regression relationships between the proportion of abundance/biomass and the proportion of sequence reads were established for each species/taxa using simple linear regression. All variables were square-root transformed to meet the assumption of homoskedasticity. In order to maximize the comparability between biomass and sequence reads, for statistically significant (p < 0.05) and relatively strong (R2 > 0.3) relationships, where the slope was less than 0.6 or greater than 1.4, the slope of the line was used to introduce linear adjustment factors for each species/taxa across the entire dataset using the equation \({n}_{\mathrm{adj}}=\frac{{s}^{2}(Tn-{n}^{2})}{T-{s}^{2}n}\), where n is the number of sequence reads of the given species/taxa, s is the slope of the regression equation, and T is the total number of sequence reads within that sample. This formula adjusts the number of sequence reads of a given taxa to change its proportion in a sample of T reads by a linear factor of s. Because changing the number of reads of one species will inevitably change the relative contribution of all other species in the sample, the adjustment factors were applied to each species or taxa in question one by one in descending order of maximum relative sequence read abundance (i.e., the taxa/species that had the greatest impact on the relative number of reads were treated first), with T re-calculated again after each adjustment. Only species/taxa that were observed in a minimum of 5 samples both via metabarcoding and microscopy were included in the analysis. The R code used to calculate and apply these conversion factors is available in Supplementary Material 2.
The proportions of sequence reads belonging to each species/taxa were then multiplied by total sample biomass (mg DW) to calculate biomass-weighed sequence reads (BWSR) for each species. Mock community analysis using abundance, biomass, and BWSR data was carried out using the package “vegan” (Oksanen et al. 2016). Additionally, in order to compare BWSR and biomass estimates directly, a “pooled” dataset was created, where each sample was represented twice, once by BWSR and once by microscopically estimated biomass data. For each dataset examined, non-metric multidimensional scaling (nMDS) was carried out on Bray–Curtis dissimilarities of fourth-root transformed data. The species significantly driving the ordination were identified using the function envfit at a significance level of p = 0.05 and visualized as biplots. Additionally, we performed hierarchical cluster analysis (average linkage method) of the resulting dissimilarity matrices, and clusters (“assemblages”) were identified via the simprof tool (Clarke et al. 2008) with an alpha level of 0.05. Finally, the correlations between Bray–Curtis dissimilarity matrices of abundance, biomass, and BWSR were examined using the Mantel permutation test (Mantel and Valand 1970), which calculates the Pearson correlation coefficient between all entries in the two matrices, while permuting the rows and columns of the matrix 9999 times to determine statistical significance. Only taxa/species that contributed a minimum of 3% of the transformed biomass, abundance, or BWSR values in at least one sample were used in the analyses. Because the relative composition of species within the samples was artificially modified, these analyses were done more as proof-of-concept for the method, rather to accurately describe the communities in this region.
Results
Sequencing summary
The sequencing run produced a total of 8,165,891 sequences, with sequences per replicate ranging between 15,000 and 182,000 reads (mean = 82,500) and per sample (across 3 replicates) between 70,700 and 457,101 (mean 250,000). The extraction blanks produced 200–400 reads and the PCR blanks 100–150 reads. There were no differences in recovered DNA concentration or sequencing depth between ethanol and dry samples, so hereafter both types of samples were used for all analyses (Fig. 2a and b). Rarefaction curves reached an asymptote in the vast majority of samples, suggesting that this sequencing depth was sufficient to recover all or most of the diversity (Supplementary Material 3).
Diversity
The visual identification yielded a total of 59 unique holoplanktonic and 16 meroplanktonic species/taxa across all samples, with the typical number of taxa per sample ranging from 15 to 40. Metabarcoding identified 357 MOTU’s across the whole dataset, of which 228 were identified to species level, 19 to genus or family level, 43 to order or class, 10 to higher taxonomic ranks, and 57 remained unassigned. Of these, 282 MOTU’s belonged to either holo- or meroplanktonic (fish or benthic) organisms and corresponded to 86 and 124 unique species/taxa of holo- and meroplankton, respectively (Supplementary Material 4). This list included all but 4 of the species identified visually (Microsetella norvegica, two appendicularian species and one monstrilloid copepod). There were no differences in species richness between ethanol and dry samples (Fig. 2c). The additional species identified via sequencing included several hard-to-identify crustaceans, hydrozoan jellyfish, and larvae of benthic animals. For example, the visually identified group Pseudo/Paracalanus spp. consisted of 5 different species found in varying proportions at different stations, of which Paracalanus parvus was the numerically dominant one.
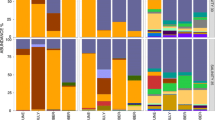
On a sample-to-sample basis, typically 90–100% of registered species/taxa were recovered via metabarcoding (average 93%), with the number of “false negatives” (species not detected via metabarcoding) never exceeding 1–4 species. However, the percentage of “false positives” (species that were identified via metabarcoding but were not detected visually, at the lowest common taxonomic resolution) was high, in many samples doubling or tripling the total diversity (Fig. 3a). When only species that contributed at least 0.01% of total reads in a sample were included, the percentage of false positives was reduced to less than 10% across all stations, but the percentage of false negatives rose, as some of the “real” species began to fall under that threshold (85% species recovered, on average) (Fig. 3c). Dropping the detection threshold further to 0.1% of total reads, false positives were reduced to no more than 1–5 species per sample, but only 60% (on average) of the “true” species on average were recovered (Fig. 3d). Regardless of the threshold limit applied, false positives typically made up a very small proportion of total reads. However, at 6 stations, they made up over 10% of total sequence reads, and at 3 stations over 25% (Fig. 4). These high contributions were typically caused by 1–2 species.
The visually observed species that were not detected via metabarcoding in all samples and/or frequently fell below the 0.01% detection limit included very small copepods such as Acartia spp., Detrichocoryceaus sp., and Oncaeidae, as well some meroplankton groups such as bivalve and gastropod larvae. Organisms frequently detected as “false positives,” on the other hand, include the large copepod Calanus hyperboreus, several species of jellyfish, fish, cirripeds, gastropods, and the chaetognath Parasagitta elegans.
Quantitative correlations
At the lowest common taxonomic level, both relative abundance and relative biomass of each species/taxa were correlated with relative sequence reads across the entire dataset, with the correlation stronger for biomass (r = 0.57) than abundance (r = 0.41). Within each individual samples, the correlations of relative biomass to sequence counts varied from 0.15 to 0.95 (Supplementary Material 5) and relative abundance to sequence counts from 0 to 0.92. When examining relative contributions of individual taxa across all samples using simple linear regression, statistically significant (p < 0.01) correlations were observed for almost all commonly encountered taxa, both between biomass and sequence counts (Fig. 5a and Table 1) and abundance and sequence counts (Table 1). The relationships to biomass were typically stronger, but for several species of copepods and pteropods, abundance was the better correlated value to relative sequence reads (Table 1). The strongest relationships to biomass were observed for several non-copepod crustacean taxa: euphausiids (R2 = 0.83), cirripeds (R2 = 0.88), ostracods (R 2 = 0.89), and decapods (R2 = 0.64), as well as echinoderm larvae (R2 = 0.74). Moderate relationships (R2 between 0.4 and 0.6) to biomass were observed for most copepod species. The weakest correlations were observed for bivalve larvae and cnidarians (R2 = 0.31). The intercept was significantly different from 0 only for Calanus helgolandicus/finmarchicus, but the slopes strongly deviated from 1 (less than 0.6 or greater than 1.4) in 22 of 40 species/taxa, indicating that they were consistently either over- or under-represented in the sequencing data relative to their estimated biomass contribution to the sample. This deviation was even stronger in the abundance data, with 29 of 40 species/taxa deviating strongly from 1. After the introduction of species- or taxa-specific linear conversion factors, all slopes fell into the range of 0.6–1.2 for biomass (Fig. 5b and Table 1). Because changing the number of reads of one species also affected the relative abundance of other species, this changed the strength of the relationship for several taxa (significance remained unchanged). For 12 species/taxa, the R2 increased by 0.05 or more, and for 5 species/taxa, it was reduced by 0.05 or more. The remainder of the relationships remained unchanged (Table 1). The overall Pearson correlation between relative biomass and sequence counts across the whole dataset improved from 0.57 to 0.77. Consistent with this high correlation, the spatial distribution of adjusted-BWSR (proportion of sequence reads multiplied by total sample biomass) showed very similar, although not identical, trends to biomass for all examined taxa (Fig. 6).
Regressions between square-root transformed proportion of biomass (estimated from microscopy) and square-root transformed proportion sequence counts for selected taxa with; a original data, and; b data adjusted with species/taxa-specific linear conversion factors. For statistical summary and all species/taxa, see Table 1
Mock community analysis
The Mantel test of associations between Bray–Curtis dissimilarity matrices of abundance and biomass, abundance and BWSR, and biomass and BWSR produced r values of 0.52, 0.54, and 0.73, respectively (significance p < 0.001), indicating moderate to high linear correlations between them. Accordingly, multivariate analysis of the three parameters (abundance, biomass, and adjusted BWSR) revealed slightly different, but highly complementary patterns for each of the datasets (Fig. 7). The separation of samples on all three nMDS ordinations were driven by 3 groups of species, with some, although not all, species shared between the data types. Overall, the metabarcoding dataset contained a much longer list of species within each of the groups due to the higher taxonomic resolution. All three ordinations contained a very distinct assemblage containing the two MultiNet stations (Group 1, light blue on Fig. 7), characterized by a variety of deep- and cold-water species that were absent in other samples. Similarly, all three datasets contained an assemblage associated with the southeast region (Group 2, pink and dark blue on Fig. 7), distinguished by Sagitta/Parasagitta spp. (all three datasets), Calanus finmarchicus/helgolandicus (biomass and BWSR), Aglantha digitale (abundance and biomass), and Candacia armata (abundance and BWSR). A northern/western group (Group 3, light and dark purple and dark green on Fig. 7) was driven by cirripeds (all three datasets, represented by 4 different species within the BWSR data), Acartia spp., (all three datasets, Acartia tonsa in the BWSR dataset), Pseudo/Paracalanus spp. (all three datasets, Pseudocalanus elongatus in the BWSR dataset), decapod larvae (abundance and BSWR, represented by 5 species in the BWSR dataset), and polychaete larvae (abundance and BSWR, represented by 4 species in the BWSR dataset). Cluster analysis of the pooled adjusted-BWSR/biomass dataset at the lowest common taxonomic resolution identified 9 clusters (“assemblages”) via the simprof routine. In 30 out of 33 samples, the microscopically derived data points were placed within the same assemblages as their BWSR counterparts. Similarly, the ethanol/dry samples collected at the same station were placed within the same assemblages in all but two instances, as was expected given the similarity of the sub-samples. The nMDS ordination partially resolved the complexity (2D stress = 0.21), and somewhat supported the separation of the clusters, although some groups were more distinct than others. Similar to the previous analysis, the most distinct assemblage included the two stations collected with the deep MultiNet (gray on Fig. 8). Despite slight variation in placement, the BWSR/biomass data points followed very similar patterns on the nMDS ordination.
Results of nMDS ordination and cluster analysis for fourth-root transformed; a abundance (estimated from microscopy); b biomass (estimated from microscopy), and c BWSR (estimated from metabarcoding) data, and the spatial distribution of clusters in the North Sea. Colors represent clusters at ~ 30–40% dissimilarity, as identified by simprof. Blue dashed lines connect samples collected at the same station. Arrows indicate biplots of species significantly (p < 0.05) correlated with the ordination, with biplots close together in space grouped as groups 1–3. For better readability, the species representing the biplots within each group are listed below the plots
Results of nMDS ordination and cluster analysis for fourth-root transformed biomass (estimated from microscopy) and BWSR data pooled at the lowest common taxonomic resolution, and the spatial distribution of clusters in the North Sea. Colors represent clusters at ~ 30–40% dissimilarity, as identified by simprof. Blue dashed lines connect samples collected at the same station, and black solid lines connect BWSR/biomass data from the same sample. Arrows indicate biplots of species significantly (p < 0.05) correlated with the ordination
Discussion
Metagenomic methods are increasingly being applied in marine ecosystem monitoring to provide deeper insights into communities of microbial organisms (Pawlowski et al. 2016; Santoferrara et al. 2020), phyto- and zooplankton (Bucklin et al. 2019; Coguiec et al. 2021; Ershova et al. 2019; Yoon et al. 2016), fish eggs and larvae (Lira et al. 2023), and zoobenthos (Klunder et al. 2022). These methods are efficient and cost-effective and offer a much higher level of taxonomic resolution compared to other approaches. However, despite increasingly coming of age, concerns remain about potential biases and errors associated with PCR-based metagenomic techniques which hinder the interpretation of the data, especially in a quantitative way. Santoferrara (2019) identified five of these potential sources of error in metabarcoding as false negatives, false positives, erroneous identifications, skewed relative abundances of different taxa, and artifactual sequences. The following sections address these potential issues in the context of our protocol and provide recommendations to address them.
Species richness detection and false positives/negatives
Metabarcoding is an extremely sensitive tool and given enough sequencing depth can detect trace amounts of DNA, including extracellular DNA, residual fragments of organisms left in the sample, and gut contents. This is an undeniable strength of the method, but this sensitivity can also vastly inflate diversity estimates, as seen in our results, where the number of “false positives” detected within a sample, at the lowest common taxonomic resolution, often was equal to or exceeded the number of species observed visually. It is important to note, however, that we cannot confirm that every “false positive” was truly absent from the sample. Although the samples in our study were to a large part artificially engineered, they still contained some individuals that we could not identify properly, such as early copepodites, nauplii, trochophore larvae, eggs, and small fragments of biological debris. For example, the presence of cnidarian or chaetognath DNA in a number of samples could have been due to these fragile gelatinous organisms breaking apart in the net haul, leaving behind fragments of their bodies. Chaetognaths are also known to regurgitate their gut contents when hauled by nets (Baier and Purcell 1997), which could result the presence of additional half-digested DNA, both their own and that of their prey. Nonetheless, regardless of whether they were truly absent or present in trace amounts, the total contribution of “false positives” was typically very low, 0.1% of total reads or lower. There were several samples, however, that fell outside of this range, and where reads belonging to false positives exceeded 5–10%, or in 3 samples even 25%, of total reads. Given the very small starting volume of the samples (300–500 individuals), it seems plausible that cross-sample contamination was a likely culprit in these cases. Alternatively, the presence of “false positive” species as early life stages, unidentifiable debris or inside the guts of other species in the sample, as described above, could potentially lead to similar results. Similar to the majority of other studies that used a mock sample approach (Santoferrara 2019), we found that metabarcoding recovered almost all of the observed diversity at the given sequencing depth, confirming that our choice of barcode and sequencing depth are appropriate for this region of the world ocean. Despite this, some of the smaller and rarer species within our data, such as the very small copepods or bivalve larvae, were not recovered in all samples, or were recovered only with a very low detection threshold, placing them within the detection range of the aforementioned “false positives.” Additionally, notably missing among the species list were appendicularians, for which COI may be a poorly suited barcode region (Bucklin et al. 2021), but which can nevertheless be significant contributors to plankton communities. Including an additional barcode, such as V4 or V9 regions of the 18S rRNA gene could have alleviated this. Other studies that implemented multiple barcode regions found that this approach increased the recovered diversity and minimized false negatives (Zhang et al. 2018). However, the numerical treatment of multi-locus metabarcoding data to estimate species relative read abundances remains a challenge.
Different approaches can be taken to minimize the effects of false positives and negatives when estimating biodiversity, depending on the goals of the study. If a broad-scale community analysis is the aim, particularly, as is often the case, in the context of earlier efforts that may have used morphological methods of identification, applying a conservative approach and setting an arbitrarily high read abundance threshold may be appropriate. Although this will exclude the rarer species, they are unlikely to be highly ecologically significant. If concurrent microscopic analysis reveals that some abundant/ecologically important species are being missed with the given metabarcoding protocol, additional genetic markers can be included, as was discussed in the section above; alternatively, these species can be screened for visually. On the other hand, if the goal is early detection of invasive species, then a very high sensitivity is necessary that will allow to detect even a single larva in a dense tissue sample. One approach to ensure proper sensitivity in this case could be “spiking” the sample with a single individual of a taxonomically similar species that does not naturally occur in the studied environment (for example, in the case of marine crustaceans, a small freshwater crustacean). This approach is commonly used in insect monitoring studies (i.e., Batovska et al. 2021). For simply recovering species richness, any number of approaches can be used. Our method of estimating diversity errs on the side of being overly conservative, counting only discreet identified species or taxonomic groups, and not MOTU’s. In most situations, not all MOTU’s can be identified to species level, but can often be identified to genus, family, or even higher ranks, and we collapsed all commonly identified MOTU’s into single categories. This removes a large part of the bias associated with clustering algorithms (i.e., van der Loos and Nijland 2021), but it is likely to underestimate true diversity, as these pooled MOTU’s can in fact represent many discreet species that are just absent from reference databases. Regardless, any kind of diversity metrics will only be relevant within the context of a single protocol and are not comparable to other methods or even other sequencing runs using similar methods, unless properly rarified. They can, however, be very valuable for within-study comparisons, for example, to map spatial, temporal, or seasonal patterns in species diversity.
Quantification and community analysis
Quantitative interpretation of metabarcoding data remains a subject of ongoing debate, with the most common concerns being difficulty of developing truly universal primers, differences in gene copy numbers between species, and the exponential nature of PCR (Bucklin et al. 2016; Santoferrara 2019; van der Loos and Nijland 2021). New, non-PCR-based molecular methods of community analysis, such as detecting specific marker genes using shot-gun sequencing, may offer a promising alternative to metabarcoding as these technologies continue to develop (Pierella Karlusich et al. 2023). Yet, despite the known issues of PCR bias, in recent years, there has been increasing evidence of quantitative or semi-quantitative potential of metabarcoding in a wide range of metazoan communities (Ershova et al. 2021; Krehenwinkel et al. 2017; Lamb et al. 2019; McLaren et al. 2019; Schenk et al. 2019; Thomas et al. 2016). One of the challenges of quantification is identifying an appropriate metric for comparing to sequence counts. With single cellular organisms, cell counts are typically used, but even then, read copy numbers can vary vastly between species (Wang et al. 2017). In metazoan communities, counts or abundance estimates are additionally confounded by the fact that different species are vastly different in size. Mesozooplankton, for example, even when captured by the same sampling gear, will vary from ca. 200 µm to several centimeters in length, which results in several orders of magnitude difference in weight. Nonetheless, we found a surprisingly high correlation of relative abundances to relative sequence reads within our dataset, particularly for some taxa. However, biomass or carbon weight was found to be the better quantitative proxy. This is not surprising, as a large organism will expectedly contain more DNA than a small one. Although species abundance is a much commonly used metric than species biomass when describing zooplankton communities, this is more a methodological artifact due to the fact that species counts are traditionally much easier to obtain than species weights and does not necessarily reflect the fact that abundance is the more ecologically relevant measure. Indeed, in terms of grazing pressure, carbon and nutrient cycling, and availability to higher trophic levels, species-specific biomass may be a much more important parameter to quantify.
Several previous studies have reported moderate to good correlations of relative read numbers to relative biomass in zooplankton for several genetic markers, including COI (Elbrecht and Leese 2015; Ershova et al. 2021; Matthews et al. 2021; Yang et al. 2017). These correlations were far better for some taxa than for others, and even the strong relationships were frequently very different from a 1: 1 ratio, which makes it challenging to infer one value from the other. We similarly found these correlations to be significant for almost all taxa for which sufficient observations were present, and likewise, observed stronger correlations for some groups than for others. We go a step further, however, and introduce a new quantitative metric called BWSR (biomass-weighed sequence reads). Unlike relative abundance, relative biomass can be much more easily transformed into a quantitative parameter by taking a single bulk biomass measurement, something that is already usually done as part of routine zooplankton analyses. This number is then multiplied by the proportion of reads belonging to a species or taxa. Furthermore, by applying species- or taxa-specific conversion factors that bring the relationships between read counts and biomass closer to unity, we can additionally reduce some of the PCR-based bias and increase comparability between these two measures. Conversion factors have been used in previous studies to improve quantification of microbial communities (McLaren et al. 2019), diatoms (Vasselon et al. 2018), fish (Thomas et al. 2016), and arthropods (Krehenwinkel et al. 2017), but to our knowledge, this is the first time this approach has been applied to marine zooplankton communities. While BWSR is closely related to biomass and shares units with it, it should nonetheless not be reported as biomass or be used to compare to other biomass-based studies. Instead, it provides a framework for quantitatively monitoring relative species composition over time and space within a single study (as for example, in Coguiec et al. 2021) and allows for the use of standard community analysis tools such as multivariate statistics. It can further be used to calculate a number of additional indicators, for example, the ratio of mero- to holoplankton, or copepod- to non-copepod taxa, or to estimate various indices of biogeographic affinity. For some taxa with a relatively small range in body weight (for example, small copepods such as Oithona spp.), BWSR values can be also used as a proxy for biomass to estimate species abundance, as is frequently done vice-versa in other studies. All three quantitative measures—abundance, biomass, and BWSR revealed very similar, and when collapsed to a common taxonomic resolution, nearly identical community patterns. The BWSR data, however, was able to provide much more detailed species-level information about which particular species were driving the community trends. For example, cirriped and decapod larvae, which were found to strongly drive the community structure in terms of all three examined parameters, were each represented by 4–5 distinct species which were not identifiable via morphology. These results strongly support the notion that metabarcoding data can be used not only for estimates of biodiversity or mapping of presence/absence of species but also to estimate relative densities of species and to quantitatively describe community structure.
Gaps and errors in reference databases
When matching sequences to reference libraries, such as BOLD (Barcode of Life System, www.boldsystems.org), three main error types are commonly encountered: (a) absence of the species in the database, (b) incorrect identification or annotation of reference vouchers, and (c) insufficient, or excessive, divergence in the genetic marker of interest that will result in either multiple species-level matches, or absence of a species-level match at the assigned threshold (usually 97%). The first error type will produce “unknowns” in the final species list, resulting in a higher proportion of “false negatives,” although frequently these MOTUs can be assigned to higher ranks (i.e., family or order). Within our dataset, only about 65% of MOTU’s were assigned to species, and 15% could not be matched even at a phylum level. The second error type, incorrect identifications, is particularly problematic because it can result in the presence of both false positives and false negatives. With the more common species, it will typically result in several species level matches in the reference database, and the investigator must decide which of them are more reliable. We encountered this type of error with one of the Acartia species, which produced a species level match for both Acartia tonsa and Acartia hudsonica. After manually checking the primary sources and annotations of these vouchers, we concluded A. hudsonica to be the more likely correct identification. On the other hand, misidentification of rare species with only a single barcoded voucher are much more difficult to recognize, and until more barcoding efforts take place, will have to be accepted on faith. Since the morphological identification of the specimen represents the quality control for the DNA barcode, ideally each barcode should be linked to a voucher specimen, and this should become standard practice going forward (Rimet et al. 2021). Multiple species level matches can also be a result of insufficient divergence in the chosen barcode to reliably discriminate between related species. Within our data, such an example was the chaetognath Eukrohnia sp., which matched to both E. hamata and E. bathyantarctica. Based on the known geographical distribution of these species, we concluded that E. hamata was the more likely correct identification. On the contrary, the chaetognath Parasagitta elegans has an exceptionally high divergence in the mitochonodrial genome, and a threshold as low as 90% similarity was used to assign this species (Marlétaz et al. 2017).
For marine metazoan zooplankton, significant efforts have been made in recent decades to create and quality control reliable DNA barcode reference libraries. Building barcode libraries and associated voucher collections have been a primary goal of several individual projects as well as national campaigns (see Bucklin et al. 2021; Weigand et al. 2019 and reference therein). Additional attempts to improve the quality of barcode libraries have also been made by implementing QA measures in reference databases (Fontes et al. 2021). A major effort in curating marine barcode data was undertaken in the Barcode of Life Data Systems (BOLD), including annotation of barcode records according to a ranking system for concordance (Radulovici et al. 2021). Moreover, a new tool for enhanced quality control of DNA barcodes of marine zooplankton (holo- and meroplankton) has been made accessible through the MetaZooGene Barcode Atlas and Database (MZGdb) (Bucklin et al. 2021). Despite the aforementioned and other ongoing efforts, available open-access DNA reference libraries remain incomplete for some taxonomic groups and/or geographical regions (McGee et al. 2019). For example, only 22% of European marine invertebrate species had at least one barcode in BOLD in 2019 (Weigand et al. 2019). Reliable barcode reference libraries are particularly important if metabarcoding is to be implemented in biomonitoring and reports of ecological status (e.g., EU Marine Strategy Framework Directive, MSFD). Thus, there is continued need to build comprehensive and reliable barcode reference databases, both by adding more taxa and by extending the geographical coverage.
Dry oven preservation as an alternative to ethanol
Fixing zooplankton samples with 80–100% ethanol is the most common way of preserving zooplankton for genetic use and is the recommended method in most barcoding or metabarcoding protocols (Rey et al. 2020; van der Loos and Nijland 2021; Wiebe et al. 2017). Other, less common, preservation methods of zooplankton include freezing of whole samples or preserving in other fixatives, such as DESS or RNAlater (Creer et al. 2016; van der Loos and Nijland 2021). We suggest another alternative for zooplankton DNA preservation, which is dehydrating whole samples in a drying oven at 65 °C for 12–24 h. In our study, we expected absolute ethanol to be the superior preservation method, but found no differences between ethanol and dry samples in recovered DNA quality or quantity, sequencing depth, or overall recovered biodiversity. There were also no differences in relative taxonomic composition, with read abundance in both preservation methods equally well correlated to relative biomass for all taxa. Although ethanol preservation remains a “non-destructive” method, allowing for subsequent sub-sampling or screening for taxa/species of interest, it comes with a set of drawbacks. These include a higher reagent cost, additional HSE risks (fumes and fire hazard), logistical transport limitations, sample storage space, and longer/more complicated processing in the lab, since most DNA extraction methods are sensitive to ethanol and require its removal. These challenges contribute to the reluctance of many larger-scale zooplankton monitoring efforts to adopt a metabarcoding component. On the other hand, many monitoring programs, like the one implemented at IMR, already collect dry samples for zooplankton biomass estimation. Simple additional steps can be taken to ensure the suitability of such samples for genetic use. This includes proper cleaning of sampling gear and lab tools between samples to prevent cross-sample contamination, prompt removal of samples from the drying oven once they are dry, and proper long-term storage. With this approach, it is recommended to keep the overall sample volume low (< 2 ml volume of wet animals with water removed) to ensure rapid and even drying. For larger samples, sub-sampling or splitting the sample onto several drying trays is recommended. The dried samples can be transported at room temperature but should be stored at − 20 °C for long-term storage. As an added benefit, measuring the dry weight of the sample prior to sequencing creates a simple path forward to convert relative sequence reads to BSWR, as described in this work.
Conclusion
In this work, we present a framework for recovering biodiversity and quantitative estimation of biomass of zooplankton species from COI metabarcoding data, which we think is suitable for cost-effective large-scale monitoring. We successfully tested the suitability and accuracy of our methods using mock zooplankton samples from the North Sea. Although some biases remain, we conclude that the methods based on COI metabarcoding have currently reached a satisfactory degree of technology readiness, and their results are comparable and highly complementary to morphology-based zooplankton analysis methods. However, similar studies should be performed in other geographical regions, for which reference barcode databases might be not so complete as for European waters. Although our results highlight the strengths of metabarcoding, they also underline the value of an integrated taxonomic approach by applying combined morphological and molecular analyses, which has also been recommended by previous studies (Di Capua et al. 2021; Matthews et al. 2021; Pierella Karlusich et al. 2022).
References
Antich A, Palacin C, Wangensteen OS, Turon X (2021) To denoise or to cluster, that is not the question: optimizing pipelines for COI metabarcoding and metaphylogeography. BMC Bioinformatics 22(1):1–24. https://doi.org/10.1186/S12859-021-04115-6/FIGURES/7
Antich A, Palacín C, San Roman D, Wangensteen O, Turon X (2019) Metabarcoding the benthic boundary layer: the role of sampling method and marker characteristics in the DNA signatures obtained at the interface between benthos and plankton. Front Mar Sci, 6. https://doi.org/10.3389/conf.fmars.2019.08.00046
Baier CT, Purcell JE (1997) Effects of sampling and preservation on apparent feeding by chaetognaths. Mar Ecol Prog Ser 146(1–3):37–42. https://doi.org/10.3354/meps146037
Batovska J, Piper AM, Valenzuela I, Cunningham JP, Blacket MJ (2021) Developing a non-destructive metabarcoding protocol for detection of pest insects in bulk trap catches. Sci Rep 11(1):1–14. https://doi.org/10.1038/s41598-021-85855-6
Boyer F, Mercier C, Bonin A, Le Bras Y, Taberlet P, Coissac E (2016) Obitools: a Unix-inspired software package for DNA metabarcoding. Mol Ecol Resour 16(1):176–182. https://doi.org/10.1111/1755-0998.12428
Bucklin A, Lindeque PK, Rodriguez-Ezpeleta N, Albaina A, Lehtiniemi M (2016) Metabarcoding of marine zooplankton: prospects, progress and pitfalls. J Plankton Res 38(3):393–400. https://doi.org/10.1093/plankt/fbw023
Bucklin A, Yeh HD, Questel JM, Richardson DE, Reese B, Copley NJ, Wiebe PH (2019) Time-series metabarcoding analysis of zooplankton diversity of the NW Atlantic continental shelf. ICES J Mar Sci 76(4):1162–1176. https://doi.org/10.1093/icesjms/fsz021
Bucklin A, Peijnenburg KTCA, Kosobokova KN, O’Brien TD, Blanco-Bercial L, Cornils A, Falkenhaug T, Hopcroft RR, Hosia A, Laakmann S, Li C, Martell L, Questel JM, Wall-Palmer D, Wang M, Wiebe PH, Weydmann-Zwolicka A (2021) Toward a global reference database of COI barcodes for marine zooplankton. Marine Biol, 168(6). https://doi.org/10.1007/s00227-021-03887-y
Clarke KR, Somerfield PJ, Gorley RN (2008) Testing of null hypotheses in exploratory community analyses: similarity profiles and biota-environment linkage. J Exp Mar Biol Ecol 366(1–2):56–69. https://doi.org/10.1016/J.JEMBE.2008.07.009
Coguiec E, Ershova EA, Daase M, Vonnahme TR, Wangensteen OS, Gradinger R, Præbel K, Berge J (2021) Seasonal variability in the zooplankton community structure in a sub-Arctic Fjord as revealed by morphological and molecular approaches. Front Mar Sci, 8. https://doi.org/10.3389/fmars.2021.705042
Creer S, Deiner K, Frey S, Porazinska D, Taberlet P, Thomas WK, Potter C, Bik HM (2016) The ecologist’s field guide to sequence-based identification of biodiversity. Methods Ecol Evol 7(9):1008–1018. https://doi.org/10.1111/2041-210X.12574
Di Capua I, Piredda R, Mazzocchi MG, Zingone A (2021) Metazoan diversity and seasonality through eDNA metabarcoding at a Mediterranean long-term ecological research site. ICES J Mar Sci 78(9):3303–3316. https://doi.org/10.1093/ICESJMS/FSAB059
Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R (2011) UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 27(16):2194–2200. https://doi.org/10.1093/bioinformatics/btr381
Elbrecht V, Leese F (2015) Can DNA-based ecosystem assessments quantify species abundance? Testing primer bias and biomass-sequence relationships with an innovative metabarcoding protocol. PLoS ONE, 10(7). https://doi.org/10.1371/journal.pone.0130324
Ershova EA, Hopcroft RR, Kosobokova KN (2015) Inter-annual variability of summer mesozooplankton communities of the western Chukchi Sea: 2004–2012. Polar Biol 38(9):1461–1481. https://doi.org/10.1007/s00300-015-1709-9
Ershova EA, Wangensteen OS, Descoteaux R, Barth-Jensen C, Præbel K (2021) Metabarcoding as a quantitative tool for estimating biodiversity and relative biomass of marine zooplankton. ICES J Mar Sci 78(9):3342–3355. https://doi.org/10.1093/ICESJMS/FSAB171
Ershova E, Descoteaux R, Wangensteen O, Iken K, Hopcroft R, Smoot C, Grebmeier JM, Bluhm BA (2019) Diversity and distribution of meroplanktonic larvae in the Pacific Arctic and connectivity with adult benthic invertebrate communities. Front Mar Sci, 6. https://doi.org/10.3389/fmars.2019.00490
Ferdous Z, Muktadir AKM (2009) A review: potentiality of zooplankton as bioindicator. Am J Appl Sci 6(10):1815–1819. https://doi.org/10.3844/AJASSP.2009.1815.1819
Fontes JT, Vieira PE, Ekrem T, Soares P, Costa FO (2021) BAGS: an automated barcode, audit & grade system for DNA barcode reference libraries. Mol Ecol Resour 21(2):573–583. https://doi.org/10.1111/1755-0998.13262
Frøslev TG, Kjøller R, Bruun HH, Ejrnæs R, Brunbjerg AK, Pietroni C, Hansen AJ (2017) Algorithm for post-clustering curation of DNA amplicon data yields reliable biodiversity estimates. Nat Commun, 8(1). https://doi.org/10.1038/s41467-017-01312-x
Gaither MR, DiBattista JD, Leray M, von der Heyden S (2022) Metabarcoding the marine environment: from single species to biogeographic patterns. Environmental DNA 4(1):3–8. https://doi.org/10.1002/EDN3.270
Geller J, Meyer C, Parker M, Hawk H (2013) Redesign of PCR primers for mitochondrial cytochrome c oxidase subunit I for marine invertebrates and application in all-taxa biotic surveys. Mol Ecol Resour 13(5):851–861. https://doi.org/10.1111/1755-0998.12138
Hirai J, Nagai S, Hidaka K (2017) Evaluation of metagenetic community analysis of planktonic copepods using Illumina MiSeq: comparisons with morphological classification and metagenetic analysis using Roche 454. PLoS ONE 12(7):e0181452. https://doi.org/10.1371/journal.pone.0181452
Klunder L, van Bleijswijk JDL, Kleine Schaars L, van der Veer HW, Luttikhuizen PC, Bijleveld AI (2022) Quantification of marine benthic communities with metabarcoding. Mol Ecol Resour 22(3):1043. https://doi.org/10.1111/1755-0998.13536
Krehenwinkel H, Wolf M, Lim JY, Rominger AJ, Simison WB, Gillespie RG (2017) Estimating and mitigating amplification bias in qualitative and quantitative arthropod metabarcoding. Sci Rep 7(1):1–12. https://doi.org/10.1038/s41598-017-17333-x
Lamb PD, Hunter E, Pinnegar JK, Creer S, Davies RG, Taylor MI (2019) How quantitative is metabarcoding: a meta-analytical approach. Mol Ecol 28(2):420–430. https://doi.org/10.1111/mec.14920
Lira NL, Tonello S, Lui RL, Traldi JB, Brandão H, Oliveira C, Blanco DR (2023) Identifying fish eggs and larvae: from classic methodologies to DNA metabarcoding. Mol Biol Rep 50(2):1713–1726. https://doi.org/10.1007/s11033-022-08091-9
Mahé F, Czech L, Stamatakis A, Quince C, De Vargas C, Dunthorn M, Rognes T (2021) Swarm v3: towards tera-scale amplicon clustering. Bioinformatics 38(1):267–269. https://doi.org/10.1093/BIOINFORMATICS/BTAB493
Mantel N, Valand RS (1970) A technique of nonparametric multivariate analysis. Biometrics 26(3):547. https://doi.org/10.2307/2529108
Marlétaz F, Le Parco Y, Liu S, Peijnenburg KTCA (2017) Extreme mitogenomic variation in natural populations of chaetognaths. Genome Biol Evol 9(6):1374–1384. https://doi.org/10.1093/gbe/evx090
Matthews SA, Goetze E, Ohman MD (2021) Recommendations for interpreting zooplankton metabarcoding and integrating molecular methods with morphological analyses. ICES J Mar Sci 78(9):3387–3396. https://doi.org/10.1093/ICESJMS/FSAB107
McGee KM, Robinson CV, Hajibabaei M (2019) Gaps in DNA-based biomonitoring across the globe. Front Ecol Evol 7:337. https://doi.org/10.3389/fevo.2019.00337
McLaren MR, Willis AD, Callahan BJ (2019) Consistent and correctable bias in metagenomic sequencing experiments. ELife, 8. https://doi.org/10.7554/eLife.46923
Møller EF, Bohr M, Kjellerup S, Maar M, Møhl M, Swalethorp R, Nielsen TG (2016) Calanus finmarchicus egg production at its northern border. J Plankton Res 38(5):1206–1214. https://doi.org/10.1093/PLANKT/FBW048
Ndah AB, Meunier CL, Kirstein IV, Göbel J, Rönn L, Boersma M (2022) A systematic study of zooplankton-based indices of marine ecological change and water quality: application to the European marine strategy framework Directive (MSFD). Ecol Indic 135:108587. https://doi.org/10.1016/J.ECOLIND.2022.108587
Oksanen J, Kindt R, Pierre L, O’Hara B, Simpson GL, Solymos P, Stevens MHHH, Wagner H, Blanchet FG, Kindt R, Legendre P, Minchin PR, O’Hara RB, Simpson GL, Solymos P, Stevens MHHH, Wagner H (2016) vegan: community ecology package, R package version 2.4–0. R Package Version 2.2–1. http://vegan.r-forge.r-project.org
Pawlowski J, Lejzerowicz F, Apotheloz-Perret-Gentil L, Visco J, Esling P (2016) Protist metabarcoding and environmental biomonitoring: time for change. Eur J Protistol 55:12–25. https://doi.org/10.1016/J.EJOP.2016.02.003
Pierella Karlusich JJ, Pelletier E, Zinger L, Lombard F, Zingone A, Colin S, Gasol JM, Dorrell RG, Henry N, Scalco E, Acinas SG, Wincker P, de Vargas C, Bowler C (2023) A robust approach to estimate relative phytoplankton cell abundances from metagenomes. Mol Ecol Resour 23(1):16–40. https://doi.org/10.1111/1755-0998.13592
Pierella Karlusich JJ, Lombard F, Irisson JO, Bowler C, Foster RA (2022) Coupling imaging and omics in plankton surveys: state-of-the-art, challenges, and future directions. Front Mar Sci, 9. https://doi.org/10.3389/fmars.2022.878803
R Development Core Team, R. (2011). R: a language and environment for statistical computing. In R foundation for statistical computing (Vol. 1, Issue 2.11.1). https://doi.org/10.1007/978-3-540-74686-7
Radulovici AE, Vieira PE, Duarte S, Teixeira MAL, Borges LMS, Deagle BE, Majaneva S, Redmond N, Schultz JA, Costa FO (2021) Revision and annotation of DNA barcode records for marine invertebrates: report of the 8th iBOL conference hackathon. Metabarcoding Metagenomics 5:207–217. https://doi.org/10.3897/mbmg.5.67862
Rey A, Corell J, Rodriguez-Ezpeleta N (2020) Metabarcoding to study zooplankton diversity. Zooplankton Ecology, 252–263. https://doi.org/10.1201/9781351021821-14
Rimet F, Aylagas E, Borja A, Bouchez A, Canino A, Chauvin C, Chonova T, Čiampor F, Costa FO, Ferrari BJD, Gastineau R, Goulon C, Gugger M, Holzmann M, Jahn R, Kahlert M, Kusber WH, Laplace-Treyture C, Leese F, … Ekrem T (2021) Metadata standards and practical guidelines for specimen and DNA curation when building barcode reference libraries for aquatic life. Metabarcoding and Metagenomics, 5, 17–33. https://doi.org/10.3897/mbmg.5.58056
Roff JC, Hopcroft RR (1986) High precision microcomputer based measuring system for ecological research. Can J Fish Aquat Sci 43(10):2044–2048. https://doi.org/10.1139/f86-251
Rognes T, Flouri T, Nichols B, Quince C, Mahé F (2016) VSEARCH: a versatile open source tool for metagenomics. PeerJ 4:e2584. https://doi.org/10.7717/peerj.2584
Santoferrara LF (2019) Current practice in plankton metabarcoding: optimization and error management. J Plankton Res 41(5):571–582. https://doi.org/10.1093/PLANKT/FBZ041
Santoferrara LF, Burki F, Filker S, Logares R, Dunthorn M, McManus GB (2020) Perspectives from ten years of protist studies by high-throughput metabarcoding. J Eukaryot Microbiol 67(5):612–622. https://doi.org/10.1111/JEU.12813
Schenk J, Geisen S, Kleinbölting N, Traunspurger W (2019) Metabarcoding data allow for reliable biomass estimates in the most abundant animals on earth. Metabarcoding Metagenomics 3:117–126. https://doi.org/10.3897/mbmg.3.46704
Thomas AC, Deagle BE, Eveson JP, Harsch CH, Trites AW (2016) Quantitative DNA metabarcoding: improved estimates of species proportional biomass using correction factors derived from control material. Mol Ecol Resour 16(3):714–726. https://doi.org/10.1111/1755-0998.12490
van der Loos LM, Nijland R (2021) Biases in bulk: DNA metabarcoding of marine communities and the methodology involved. Mol Ecol 30(13):3270–3288. https://doi.org/10.1111/mec.15592
Vasselon V, Bouchez A, Rimet F, Jacquet S, Trobajo R, Corniquel M, Tapolczai K, Domaizon I (2018) Avoiding quantification bias in metabarcoding: application of a cell biovolume correction factor in diatom molecular biomonitoring. Methods Ecol Evol 9(4):1060–1069. https://doi.org/10.1111/2041-210X.12960
Wang C, Zhang T, Wang Y, Katz LA, Gao F, Song W (2017). Disentangling sources of variation in SSU rDNA sequences from single cell analyses of ciliates: impact of copy number variation and experimental error. Proceedings of the Royal Society B: Biological Sciences, 284(1859). https://doi.org/10.1098/RSPB.2017.0425
Wangensteen OS, Palacín C, Guardiola M, Turon X (2018) DNA metabarcoding of littoral hardbottom communities: high diversity and database gaps revealed by two molecular markers. PeerJ, 2018(5). https://doi.org/10.7717/peerj.4705
Weigand H, Beermann AJ, Čiampor F, Costa FO, Csabai Z, Duarte S, Geiger MF, Grabowski M, Rimet F, Rulik B, Strand M, Szucsich N, Weigand AM, Willassen E, Wyler SA, Bouchez A, Borja A, Čiamporová-Zaťovičová Z, Ferreira S, … Ekrem T (2019). DNA barcode reference libraries for the monitoring of aquatic biota in Europe: gap-analysis and recommendations for future work. Sci Total Environ, 678, 499–524. https://doi.org/10.1016/J.SCITOTENV.2019.04.247
Wiebe PH, Bucklin A, Benfield M (2017) Sampling, preservation and counting of samples: II. Zooplankton. In Marine plankton: a practical guide to ecology, methodology, and taxonomy (Vol. 1, pp. 104–136). Oxford University Press. https://doi.org/10.1093/OSO/9780199233267.003.0010
Yang J, Zhang X, Xie Y, Song C, Zhang Y, Yu H, Burton GA (2017) Zooplankton community profiling in a eutrophic freshwater ecosystem-lake tai basin by DNA metabarcoding. Sci Rep, 7(1). https://doi.org/10.1038/s41598-017-01808-y
Yang J, Zhang X (2020) eDNA metabarcoding in zooplankton improves the ecological status assessment of aquatic ecosystems. Environ Int 134:105230. https://doi.org/10.1016/J.ENVINT.2019.105230
Yoon TH, Kang HE, Kang CK, Lee SH, Ahn DH, Park H, Kim HW (2016) Development of a cost-effective metabarcoding strategy for analysis of the marine phytoplankton community. PeerJ 2016(6):e2115. https://doi.org/10.7717/PEERJ.2115/SUPP-1
Zhang GK, Chain FJJ, Abbott CL, Cristescu ME (2018) Metabarcoding using multiplexed markers increases species detection in complex zooplankton communities. Evol Appl 11(10):1901–1914. https://doi.org/10.1111/eva.12694
Acknowledgements
We thank Gayantonia Franzè, the North Sea Ecosystem program, and the captain and crew of the R/V Johan Hjort for providing cruise time and logistical support to collect and prepare the samples. We also thank Magnus Reeve and Gastón Aguirre for help with sample collection, processing, and specimen identification. We sincerely acknowledge Hanne Sannæs, Karen Martinez-Swatzon, and Ida Mellerud at the IMR Flødevigen Station for providing a warm and welcoming environment at the molecular laboratory facility and offering technical support at every step of the way. OW is a member of the research group 2021 SGR 01271 funded by the Generalitat de Catalunya (AGAUR).
Funding
Open access funding provided by Institute Of Marine Research This work was financially supported by the Research Council of Norway (Norges forskningsråd) through the CoastRisk initiative (NFR 299554/F40).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflict of interest.
Ethical approval
No animal testing was performed during this study.
Sampling and field studies
All necessary permits for sampling and observational field studies have been obtained by the authors from the competent authorities and are mentioned in the acknowledgements, if applicable.
Data availability
The original data used to produce this manuscript can be downloaded from the Norwegian Marine Data Centre (NMDC) at https://doi.org/10.21335/NMDC-148996217. The raw sequencing data can be found at NCBI under BioProject PRJNA947953.
Author contribution
EE and TF conceived and designed the study. EE carried out the sample collection, sample analysis, and molecular laboratory work. EE and OW did the bioinformatics. EE analyzed the data and wrote the manuscript with contributions from OW and TF. All authors have read and approved the manuscript.
Additional information
Communicated by A. Weydmann-Zwolicka
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
This article is a contribution to the Topical Collection Discovering the water column: integrated taxonomic approaches for measuring marine pelagic biodiversity
Supplementary information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Ershova, E.A., Wangensteen, O.S. & Falkenhaug, T. Mock samples resolve biases in diversity estimates and quantitative interpretation of zooplankton metabarcoding data. Mar. Biodivers. 53, 66 (2023). https://doi.org/10.1007/s12526-023-01372-x
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s12526-023-01372-x