Abstract
Although Ipnops specimens are relatively common in the Clarion-Clipperton Fracture Zone (CCZ), an area targeted for potential future deep-sea mining, a reliable species identification has not yet been possible due to the lack of a captured specimen. In April 2012, an Ipnops specimen was caught for the first time from the eastern CCZ during an exploration cruise of the BGR. Species identification of this specimen was performed using a comparative application of morphological analysis and DNA barcoding and resulted in its clear assignment to Ipnops meadi Nielsen, 1966. Of the 23 compared morphological characters, 22 are inside the ranges available for I. meadi. Molecular analyses show a sequence distance of 0.76% divergence to an Ipnops specimen collected off Hawaii, close to the CCZ and also within the known geographical distribution range of I. meadi. The additional study of five specimens of I. meadi from the Arabian Sea has extended the previously known range of the following morphological characters of this species: gill rakers on anterior arch (17–21), head length (17.6–24.0 % SL), upper jaw length (10.7–14.0 % SL), maximum width of eye-plates (7.8–9.8 % SL), preanal length (58.8–79.0 % SL), and predorsal length (34.5–40.5 % SL). Ipnops specimens deriving from Australian waters could not be clearly assigned with confidence to one of the valid Ipnops species based on current morphological and molecular analyses. It seems possible that at least one previously undescribed Ipnops species occurs in Australian waters and further work is required on the genus to resolve uncertainties.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The Clarion Clipperton Fracture Zone (CCZ) is an area of great interest as it holds the largest known deposits of polymetallic nodules, which are rich in metals such as manganese, nickel, cobalt and copper and are being targeted for potential mining (Miller et al. 2018; Hein et al. 2020). Mining of polymetallic nodules has been prompted by the demand for metals necessary for high technology applications and renewable energy infrastructure (Hein et al. 2020). Despite the growing interest as mirrored in a substantial number of recent scientific expeditions, baseline information on the associated megafaunal communities and their ecology is still restricted and based largely on imagery studies (e.g. Harbour et al. 2020; Simon-Lledó et al. 2020; Drazen et al. 2021). Although the importance of these studies is unquestionable, there is a need for specimen collection and thus verified species identifications necessary for future management and conservation actions (Amon et al. 2016; Amon et al. 2017; Bribiesca-Contreras et al. 2022; Christodoulou et al. 2022).
It has been assumed in several studies, based on seabed imagery, that the relatively common Ipnops specimens in the CCZ could be assigned to the species Ipnops meadi Nielsen, 1966 (e.g. Amon et al. 2017; Drazen et al. 2021). Okiyama and Ida (2010) also assumed that the Ipnops specimen photographed by them in the Japan Trench, off Cape Erimo, was I. meadi. However, a reliable species identification has not yet been possible due to a lack of a captured specimen. During the BIONOD exploration cruise of the German Federal Institute for Geosciences and Natural Resources (BGR) in the Eastern Central Pacific Ocean, in April 2012, a specimen belonging to the genus Ipnops (Aulopiformes: Ipnopidae) was caught for the first time in the CCZ.
Fishes of the aulopiform family Ipnopidae number 33 species in six genera (Nelson et al. 2016; Fricke et al. 2022) and occur in tropical and temperate waters of the Atlantic, Indian and Pacific Oceans at depths between 476 and 6000 m (McEachran and Fechhelm 1998; Franco et al. 2009).
The species are mostly small and slender with a depressed head and flattened abdomen (Nielsen 1966). The mouth is large, reaching far behind the eye, the slightly protruding lower jaw has a fleshy tip, the teeth are small and needle-shaped, the gill slits are wide and the gill rakers are long (Paxton and Niem 1999; Bray 2017). They have a lateral line along the midline of the body, deciduous scales, a short-based dorsal fin located anterior to the anal fin and some species have elongated pectoral, pelvic and caudal rays (Bray 2017).
The species of the ipnopid genus Ipnops Günther, 1878 have dorsally directed, flat and degenerated eyes without a lens on the upper surface of the head covered by transparent frontals and parietals forming a thin bony membrane (Nielsen 1966). The genus Ipnops comprises the following four species (Fricke et al. 2022): I. agassizii Garman, 1899, I. meadi Nielsen, 1966, I. murrayi Günther, 1878 and I. pristibrachium (Fowler, 1943).
Although the horizontal and vertical distributions of these species in the world’s oceans are not yet fully known, some differences do exist. Ipnops agassizii is distributed circumglobally in warmer temperate waters of the Atlantic, Indian and Pacific Oceans. In the Eastern Central Atlantic Ocean, this species occurs east of the mid-Atlantic Ridge between 5 and 6°N, as well as off Namibia, Mauritania and Cape Verde (Bannerman et al. 2015). Its known depth distribution ranges from 1392 to at least 2820 m (Nielsen 1966). Ipnops meadi is known to occur in the Indo-Pacific Ocean off Kenya, the Seychelles, the Maldives, Sri Lanka, northern and southern Japan, Indonesia (Sulawesi), the Magellan Rise near Hawaii in the Eastern Central Pacific Ocean and off Peru (Russell et al. 2020). This species was found at depths between 3310 and 4940 m (Nielsen 1966). Ipnops murrayi occurs in the Atlantic Ocean from the Bahamas to the Gulf of Mexico and Caribbean Sea. It has also been recorded from southeastern Brazil and Tristan da Cunha, whereas specimens from the Cape Verde Islands and Mauritania require verification (Moore and Polanco Fernandez 2019). Its known depth range is 1555–3475 m (Nielsen 1966). Ipnops pristibrachium is only known from off Sulawesi in the Western Central Pacific Ocean (Holleman et al. 2020) from a depth of 1525–1992 m (Nielsen 1966). This species was once considered a synonym of I. agassizii (Sulak 1990), but was recently considered as a valid species (Chen 2002; Fricke et al. 2022).
This work aims to present a detailed species identification of the first Ipnops specimen caught in the CCZ based on the integration of morphological characters and DNA barcodes.
Material and methods
Specimen sampling and processing
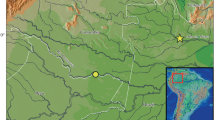
An Ipnops specimen was caught in the CCZ, Eastern Central Pacific Ocean, during the BIONOD cruise of BGR with RV L’Atalante on the 2nd of April 2012. The specimen was sampled from just (< 2 m) above the seafloor using an epibenthic sledge (station 06EBS) whilst trawling from 11°42.76′N, 116°40.35′W (4261 m depth) to 11°46.22′N, 116°41.13′W (4259 m depth). The catch location of the specimen is shown in Fig. 1.
a Bathymetric map (GEBCO 2014, International Seabed Authority 2020, 2021) showing the catch location (black star) of the specimen of Ipnops meadi Nielsen, 1966 obtained from the German Contract Area, CCZ (marked in grey), in the Eastern Central Pacific; b bathymetric map showing 42 locations in the German Contract Area at which Ipnops specimens were recorded based on available seafloor images
Immediately after collection, the specimen was preserved in pre-cooled 96% ethanol and a tissue sample was taken for molecular analysis at Senckenberg’s German Center for Marine Biodiversity Research (DZMB, Wilhelmshaven, Germany). About 8 years later, the specimen was transferred to the Zoological Museum Hamburg (ZMH), where it was deposited in the fish collection under catalogue number ZMH 25593.
Eighteen Australian specimens were collected from the RV Investigator with a beam trawl (4 m wide, 0.5 m high, mouth area 2 m2) designed to sample fishes and invertebrates from flat to low relief seafloor habitats (Lewis 2010). The latitude and longitude data for the stations are for the start of the trawl and are listed in the “Morphological comparisons” section.
A small piece of muscle tissue (for DNA analysis) was extracted from Australian specimens collected in 2015 and 2017 and subsequently frozen, before whole fishes were fixed in 10% formaldehyde and deposited in the CSIRO Australian National Fish Collection (ANFC), Hobart. ANFC registration numbers are listed below in the “Morphological comparisons” section and in Table 2.
Image transect data
Occurrence data of Ipnops in the German Contract Area in the CCZ were collected using photographic images obtained with towed camera systems. Images were obtained from transects during the cruises BIONOD on RV L’Atalante (March 29–May 10, 2012), MANGAN14 on R/V Kilo Moana (April 15–June 03, 2014), FLUM on RV Sonne (May 03–June 16, 2015), and MANGAN18 on RV Sonne (April 05–May 29, 2018). All transects were annotated using the annotation platform BIIGLE (Langenkämper et al. 2017) as set up on the server of the Senckenberg Nature Research Society. A total of 42 occurrences of Ipnops specimens were recorded using these photographic images (for locations, see Fig. 1). Three selected photographs of Ipnops specimens obtained with towed camera systems are available in Online Resource 1.
Morphological analyses
Meristic counts (Table 1) were made on the left-hand side of the specimens according to Hubbs and Lagler (1958) with a modification concerning the urostyle that was not included in the vertebral counts according to Nielsen (1966). Vertebrae; the rays of dorsal, anal and caudal fins; and other osteological elements were examined from radiographs using an X-ray imaging system (Faxitron LX-60). Digital radiographs of Australian specimens were created using an Inspex 20i 70kVp Microfocus source with LTX-1717 X-ray Detector (Kodex Inc.). External morphometric measurements were taken by vernier caliper to one tenth of a millimetre following Hubbs and Lagler (1958) with the exception that the symphysis of the upper jaw was used as the anteriormost part instead of the protruding lower jaw as in Nielsen (1966). All morphometric characters are provided as a percentage of the standard length. Images of the specimen were taken with a Digital BK Plus imaging system (Dun, Inc.), equipped with a Canon EOS 5DS DSRL camera with a 100-mm macro lens. Image stacking was performed using Zerene Stacker v.1.04 (Zerene Systems LLC.).
Morphological comparisons
We have compared the Ipnops specimen from the CCZ with all four currently known Ipnops species based on their morphological characters provided by Nielsen (1966) and Franco et al. (2009). Furthermore, the following additional material was analysed for morphological comparisons: I. meadi: ZMH 25834, 4 specimens, 84–112 mm SL, Northwestern Indian Ocean, Arabian Sea, 14°30′N, 64°38′E, RV Meteor, cruise 33/1, station 647, otter trawl, 3940 m depth, 11 October 1995. ZMH 25835, 1 specimen, 113 mm SL, Northwestern Indian Ocean, Arabian Sea, 14°26′N, 64°32′E, RV Meteor, cruise 33/1, station 649, otter trawl, 3953 m depth, 12 October 1995. Ipnops sp. 1 (Eastern Indian Ocean, Great Australian Bight, South Australia, all collected by RV Investigator): CSIRO H 7906-01, 1 specimen, 130 mm SL, 34°04.44′S, 129°10.92′E, IN2015_C01/064, 2649–2803 m depth, 13 November 2015. CSIRO H 7920-04, 1 specimen, 137 mm SL, 35°49.10′S, 134°06.54′E, IN2015_C02/141, 2852–2800 m depth, 05 December 2015. CSIRO H 7933-03, 1 specimen, 129 mm SL, 35°00.56′S, 130°19.02′E, IN2015_C02/227, 2848–2831 m depth, 11 December 2015. CSIRO H 8091-05, 1 specimen, 125 mm SL, 35°48.89′S, 132°01.27′E, IN2017_C01/175, 3930–4250 m depth, 15 April 2017. CSIRO H 8092-01, 1 specimen, 114 mm SL, CSIRO H 8092-08, 1 specimen, 56 mm SL, and CSIRO H 8092-11, 1 specimen, 131 mm SL, 35°42.95′S, 131°39.38′E, IN2017_C01/178, 3817–3950 m depth, 16 April 2017. CSIRO H 8096-04, 1 specimen, 133 mm SL, 34°26.84′S, 129°31.90′E, IN2017_C01/197, 3235–3350 m depth, 21 April 2017. CSIRO H 8097-03, 1 specimen, 120 mm SL and CSIRO H 8097-05, 1 specimen, 106 mm SL, 34°32.92′S, 129°36.12′E, IN2017_C01/198, 3389–3540 m depth, 21 April 2017.
Ipnops sp. 2 (Southwestern Pacific Ocean, Eastern Australia, all collected by RV Investigator): CSIRO H 8114-01, 1 specimen, 140 mm SL (largest of 2 specimens in lot), NE of Flinders Island, Bass Strait, Tasmania, 39°27.72′S, 149°16.56′E, IN2017_V03/022, 2760–2692 m depth, 22 May 2017. CSIRO H 8118-03, 1 specimen, 140 mm SL, E of Bermagui, New South Wales, 36°21.30′S, 150°38.64′E, IN2017_V03/044, 2821–2687 m depth, 27 May 2017. CSIRO H 8119-01, 1 specimen, 138 mm SL, SE of Jervis Bay, New South Wales, 35°19.98′S, 151°15.48′E, IN2017_V03/056, 2650–2636 m depth, 29 May 2017. CSIRO H 8120-04, 1 specimen, 136 mm SL (largest of 3 specimens in lot), E of Newcastle, New South Wales, 32°58.10′S, 152°57.12′E, IN2017_V03/067, 2704–2902 m depth, 31 May 2017. CSIRO H 8127-01, 1 specimen, 128 mm SL, Central Eastern Commonwealth Marine Reserve, New South Wales, 30°05.86′S, 153°53.92′E, IN2017_V03/086, 2429–2518 m depth, 05 June 2017. CSIRO H 8128-01, 1 specimen, 114 mm SL, E of Byron Bay, New South Wales, 28°40.59′S, 154°12.20′E, IN2017_V03/090, 2587–2562 m depth, 07 June 2017. CSIRO H 8131-01, 1 specimen, c. 136 mm SL (genetic data only as specimen not found at CSIRO), E of Moreton Bay, Queensland, 26°56.75′S, 153°56.70′E, IN2017_V03/101, 2520–2576 m depth, 09 June 2017. CSIRO H 8138-02, 1 specimen, 96 mm SL, Coral Sea Commonwealth Marine Reserve, Queensland, 23°45.06′S, 154°38.34′E, IN2017_V03/122, 2369–2329 m depth, 13 June 2017.
Comparison of the main characters is given in Table 1, where differences between the Ipnops specimen from the CCZ and the other Ipnops material are indicated in bold.
DNA extraction, amplification and sequencing
Total genomic DNA from the Ipnops specimen from the CCZ was extracted at the DZMB using the Qiagen DNeasy Blood and Tissue Kit following the manufacturers’ protocol. A 657-bp fragment of the mitochondrial (mt) cytochrome oxidase sub-unit I (COI) gene was amplified by polymerase chain reaction (PCR). Amplifications were performed using AccuStart PCR SuperMix (ThermoFisher Scientific) in a 25-μL volume containing 12.5 μL AccuStart PCR SuperMix, 9.5 μL ddH2O, 0.5 μL of each primer (10 pmol μL−1) and 2 μL of DNA template. For the COI amplification, a primer cocktail (C_FishF1t1-C_FishR1t1) including FishF2_t1 (5′-TGTAAAACGACGGCCAGTCGACTAATCATAAAGATATCGGCAC), FishR2_t1 (5′-CAGGAAACAGCT ATGACACTTCAGGGTGACCGAAGAATCAGAA), VF2_t1 (5′-TGTAAAACGACGGCCAGTCAACCAACC ACAAAGACATTGGCAC) and FR1d_t1 (5′-CAGGAAACAGCTATGACACCTCAGGGTGTCCGAARAAYCA RAA) (Ivanova et al. 2007) tailed with M13F and M13R-pUC was used. The amplification conditions consisted of an initial denaturation step of 3 min at 94°C, 35 cycles of 30 s at 94°C, 60 s at 47°C and 1 min at 72°C, followed by a final extension step of 5 min at 72°C. All PCR products were purified using ExoSap-IT (ThermoFisher Scientific). Amplified fragments were sequenced in both directions at Macrogen Europe Laboratory (Amsterdam, The Netherlands). Forward and reverse sequences were assembled and edited using Geneious v.9.1.7 (https://www.geneious.com; Kearse et al. 2012).
DNAs from muscle samples from the Australian specimens were extracted using the Wizard® SV Genomic DNA Purification system (Promega, Australia) with starting material of 0.25g. Tissue extractions were undertaken using SV mini-columns following the manufacturer’s instructions (including an overnight digestion at 55°C) and the addition of 20μL Proteinase K (20mg/mL, Promega). DNAs were individually precipitated in 160μL nuclease-free water. DNA was quantified on a Nanodrop 8000 UV-Vis Spectrophotometer (Thermo Scientific, USA) and aliquoted into 96-well plates. The DNA samples were sent at room temperature to the Ramaciotti Centre for Genomics (UNSW Sydney, Australia) where a portion of the COI was amplified (using FishF1&F2 and FishR2 primers (Ward et al. 2005) and an annealing temperature of 54°C). Bi-directional cycle sequencing was then undertaken using the abovementioned PCR primers and BigDye® Terminator v3.1 Cycle sequencing kit (Life Technologies, USA) on the PCR products, with cycle sequenced products run on an ABI3730XL Autosequencer (Applied Biosystems, USA) at Ramaciotti. Raw forward and reverse sequences were de novo assembled in Geneious v8.1.9 (https://www.geneious.com). Remaining archival DNA is stored at −80°C at the CSIRO Marine Laboratories in Hobart.
Processing of COI sequences
The obtained COI sequences were compared with the GenBank nucleotide database using BLASTN (Altschul et al. 1990). Our dataset consists of the new Ipnops sequence, the 18 sequences from the ANFC and four additional Ipnops sequences from either BOLD or GenBank. The COI sequences were aligned using MAFFT v7.308 under G-INS-I algorithm (Katoh et al. 2002). Sequence divergences were estimated as uncorrected p-distances using MEGA7 (Table 2). A neighbour-joining tree was constructed in MEGA7 using a p-distance substitution model, treating gaps and missing data with “pairwise deletion” and by running 1000 bootstrap replicates (Fig. 3).
Specimen metadata and sequences were uploaded to the Barcode of Life Data System. Additionally GenBank Accession numbers were acquired and are listed in Table 2, which are available for all recorded specimen sequences except for one specimen from the Caribbean Sea.
Results and discussion
Morphological description of the specimen
Counts and measurements of the collected CCZ specimen (ZMH 25593) are provided in Table 1. The specimen belongs to the genus Ipnops according to the characters given by Nielsen (1966). The morphological description of the specimen is as follows:
Body long and slender, its abdominal part flattened. Anus under ventral fins, just anterior to dorsal fin. Scales relatively large, body and sides of the head scaled (Fig. 2). Scales absent on fins, except for the caudal fin, whose base is partly covered by scales.
Ipnops meadi, ZMH 25593, 124 mm SL, captured in the CCZ by epibenthic sledge during the BIONOD cruise of BGR on the 2nd of April 2012 (trawling from 11°42.76′N, 116°40.35′W to 11°46.22′N, 116°41.13′W; black star in Fig. 1). Top: picture of preserved specimen; bottom: radiograph of preserved specimen. Important meristic characters for species identification have been labelled (also see Table 1)
Head depressed dorsoventrally; its width greater than its height. Eye-plates on the upper surface of the head, dorsally directed, flat and without a lens; eyes covered with a thin and transparent bony membrane formed by frontals and parietals. Otoliths present (Fig. 2).
Mouth large. Lower jaw slightly protruding with relatively large pores. Anterior three pores smaller than the distance between pores, posterior three pores equal to or larger than the distance between pores. Upper jaw accounts for approximately 58 % of the head length. Gill slits long. 18 long and relatively thin gill rakers on the anterior arch, three on the upper branch, one on the angle and 14 on the lower branch.
Dorsal fin short-based (7.4 % SL) and located in front of anal fin. Anterior ray of dorsal fin placed over the 18th vertebra (Table 1) and much closer to snout than to the base of caudal fin (predorsal length 36.9 % SL). Origin of anal fin placed below the 36th vertebra and closer to caudal base than to the anus (preanal length 67.6 % SL). Pelvic fin base far anterior of dorsal fin. Dorsal ray of pectoral fin relatively long.
Teeth on jaws fine and numerous, only a few small pointed teeth on vomer and palatine. A lateral line present along the midline of the body (Fig. 2). Head, neck, chest, mouth and gill cavity are dark-brown after 10 years of preservation in 96% ethanol. Body yellow-brown, fins lighter.
Integrative taxonomic identification of Ipnops meadi Nielsen, 1966
ZMH 25593 differs clearly from I. murrayi in the following characteristics: fewer gill rakers (18 vs. 20–23) and lateral line scales (51 vs. 53–58), shorter upper jaw length (10.8 vs. 11.5–13.5), shorter maximum width of eye-plates (7.5 vs. 8.3–10.0), shorter length of dorsal (7.4 vs. 8.3–13.1) and anal fin bases (14.4 vs. 16.9–20.0), longer predorsal length (36.9 vs. 30.7–36.8), and longer upper pectoral rays (Table 1).
ZMH 25593 is clearly distinguished from its other two congeners, I. agassizii and I. pristibrachium, by a number of key characters. In total, out of 23 characters compared, ZMH 25593 differs in 15 characters from I. agassizii and in 14 characters from I. pristibrachium (for details, see Table 1). Therefore, an assignment of ZMH 25593 to these species can be excluded.
The characteristics of the Ipnops specimen from the CCZ agree well with the counts and measurements given by Nielsen (1966) for I. meadi from the Western Indian Ocean to the Eastern Central Pacific Ocean and of I. meadi from the Arabian Sea (Table 1). All counts fall within the minimum and maximum values of all 13 meristic characters of the 26 analysed specimens of I. meadi. Nine out of ten measurements are also within the ranges available for I. meadi (Table 1). Only the maximum width of eye-plates is slightly smaller than the known ranges for I. meadi. This difference can perhaps be attributed to the relatively small number of I. meadi specimens (only 26) which have been morphologically analysed in detail to date and to the fact that ZMH 25593 is the largest specimen of I. meadi studied so far.
The additional study of five specimens of I. meadi from the Arabian Sea (ZMH 25834 and ZMH 25835) has somewhat extended the range of the following characters compared to Nielsen (1966): gill rakers on anterior arch (17–21), head length (17.6–24.0 % SL), upper jaw length (10.7–14.0 % SL), maximum width of eye-plates (7.8–9.8 % SL), preanal length (58.8–79.0 % SL), and predorsal length (34.5–40.5 % SL).
It should also be mentioned here that Nielsen (1966) stated the absence of otoliths as a feature of I. meadi in contrast to all other Ipnops species. Marshall and Staiger (1975) reported that this could be the result of long preservation periods in formaldehyde rather than an actual character of the species. J. G. Nielsen himself confirmed this later (Franco et al. 2009). This explains the presence of otoliths in ZMH 25593, as this specimen was not preserved in formaldehyde.
It should be noted here that based on our study, we cannot confirm the current view that I. pristibrachium is a valid species (Chen 2002, Fricke et al. 2022), because almost all of its characters are within the range of I. agassizii (Table 1). In this aspect, we rather follow Nielsen (1966) that I. pristibrachium should be considered a synonym of I. agassizii.
ZMH 25593 is also clearly distinguishable from the Australian Ipnops specimens not yet assigned to a species. It differs in 12 characters from Ipnops sp. 1 from the Great Australian Bight and in 14 characters from Ipnops sp. 2 from East Australia (for details, see Table 1).
The results of the molecular analyses agree well with the morphological analyses. The COI sequence (ON526742) of I. meadi collected from the CCZ is very similar (p-distance: 0.76%, Table 2; Fig. 3) to a COI sequence of an Ipnops specimen, identified as Ipnops cf. meadi from the Eastern Central Pacific Ocean (off Hawaii). Both specimens are within the known distribution area of I. meadi and in close proximity to each other (Table 2). This is the first time a verified COI sequence is available for this species.
Neighbour-joining tree of p-distance of Ipnops Günther, 1878 based on COI sequences. Numbers on nodes indicate neighbour-joining bootstrap values. Only values above 95% are shown
Much higher mean genetic distances were found when compared with I. murrayi (13.52–13.67%) from the Western Central Atlantic Ocean, Ipnops sp. 1 from the Great Australian Bight (GAB) (13.65–14.42%) and Ipnops sp. 2 from East Australia (14.09–15.16%).
However, the Ipnops specimens from Australian waters could not be clearly assigned to one of the valid Ipnops species based on the molecular and morphological analyses. The morphological data for the two genetic lineages of Australian Ipnops do not unambiguously align with any one species of Ipnops from available data. The GAB samples match Atlantic samples identified as ‘I. murrayi’ using the BOLD identification tool (not depicted in the tree as not publicly available). Nielsen (1966) discussed two specimens from the Western Indian Ocean that did not conform to any Ipnops species. He postulated that due to the specimens having characters in common with all three species of Ipnops that they could be hybrids and even raised the possibility of the species being subspecies (Nielsen 1966). Large COI sequence divergences (>5%) between multiple Australian forms points to further molecular and morphological investigations being necessary to resolve any possibly undescribed Ipnops species.
Final discussion and conclusion
Abyssal communities in the CCZ will be affected by potential future deep-sea mining activities. In order to evaluate the potential impact of nodule removal, contractors are required to comprehensively assess the biodiversity in their contract areas prior to any mining activity. The demersal fish fauna is an important component of the abyssal communities. Most of the records of reported fish species are based on images collected by remotely operated vehicles (ROV), towed cameras or baited camera landers (e.g. Harbour et al. 2020; Drazen et al. 2021; Leitner et al. 2021). None of these devices is able to collect specimens for accurate species identification. Baited traps used to collect scavengers (primarily amphipods) do potentially collect some fishes (mostly species of Macrouridae, Ophidiidae and Zoarcidae; Drazen et al. 2021), but Ipnops has not been attracted by these devices so far. Ipnops is the most common fish genus observed in transects in the CCZ according to presently available abyssal imagery (Drazen et al. 2021). The distinctive reflection of light from its unique plate-like eyes (Online Resource 1) present in dorsal position makes the genus Ipnops easy to identify and detect. However, for species identification, voucher specimens are required. With this contribution, we report on the first collection of an Ipnops specimen from the CCZ. We have been able to match the morphological features with a DNA barcode retrieved from the COI gene. We can now confirm that the specimen reported from Hawaii also belongs to I. meadi and can exclude I. murray, I. agassizii and I. pristibrachium as potential species. The correct identification of the species present in the CCZ is crucial for the assessment of the potential impact of mining. In the present case, the confirmed range of distribution of I. meadi from the Indian Ocean to the Eastern Pacific Ocean makes it unlikely that deep-sea mining will threaten the species at a global scale, but it is reasonable to assume that they will be affected in mining areas, perhaps reducing their local standing stocks. However, this identification of a single voucher specimen does not necessarily conclude the existence of only one Ipnops species in the area. Very little is known on genetic diversity and connectivity of demersal fish populations at large geographic scales. This contribution is a first step in understanding distributional ranges in the abyss by matching taxonomy with morphological and genetic information for a common fish species in a potential mining area.
References
Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215:403–410. https://doi.org/10.1016/S0022-2836(05)80360-2
Amon DJ, Ziegler AF, Dahlgren TG, Glover AG, Goineau A, Gooday AJ, Wiklund H, Smith CR (2016) Insights into the abundance and diversity of abyssal megafauna in a polymetallic-nodule region in the eastern Clarion-Clipperton Zone. Sci Rep 6:30492. https://doi.org/10.1038/srep30492
Amon DJ, Ziegler A, Drazen J, Grischenko A, Leitner A, Lindsay D, Voight J, Wicksten M, Young C, Smith C (2017) Megafauna of the UKSRL exploration contract area and eastern Clarion-Clipperton Zone in the Pacific Ocean: Annelida, Arthropoda, Bryozoa, Chordata, Ctenophora. Mollusca Biodivers Data J 5:e14598. https://doi.org/10.3897/BDJ.5.e14598
Bannerman P, Nunoo F, Poss S, Russel B (2015) Ipnops agassizii. The IUCN Red List of Threatened Species 2015: e.T13462777A15603215. https://doi.org/10.2305/IUCN.UK.2015-4.RLTS.T13462777A15603215.en. Accessed 11 March 2022
Bray DJ (2017) Spiderfishes, Ipnopidae. In: Fishes of Australia. https://fishesofaustralia.net.au/home/family/46. Accessed 12 Mar 2022
Bribiesca-Contreras G, Dahlgren TG, Amon DJ, Cairns S, Drennan R, Durden JM, Eléaume MP, Hosie AM, Kremenetskaia A, McQuaid K, O’Hara TD, Rabone M, Simon-Lledó E, Smith CR, Watling L, Wiklund H, Glover AG (2022) Benthic megafauna of the western Clarion-Clipperton Zone, Pacific Ocean. Zookeys 1113:1–110 https://zookeys.pensoft.net/article/82172/
Chen S-Z (2002) Fauna Sinica. Osteichthyes. Myctophiformes, Cetomimiformes, Osteoglossiformes. Science Press, Beijing: i-ix + 1-349
Christodoulou M, De Grave S, Vink Α, Martinez Arbizu P (2022) Taxonomic assessment of deep-sea decapod crustaceans collected from polymetallic nodule fields of the East Pacific Ocean using an integrative approach. Mar Biodivers 52:61. https://doi.org/10.1007/s12526-022-01284-2
Drazen JC, Leitner A, Jones DOB, Simon- Lledó E (2021) Regional variation in communities of demersal fishes and scavengers across the CCZ and Pacific Ocean. Front Mar Sci 8:630616. https://doi.org/10.3389/fmars.2021.630616
Fowler HW (1943) Contributions to the biology of the Philippine Archipelago and adjacent regions. Bulletin of the United States National Museum no. 100, v. 14, 2: 53-91
Franco MAL, Braga AC, Nunan GWA, Costa PAS (2009) Fishes of the family Ipnopidae (Teleostei: Aulopiformes) collected on the Brazilian continental slope between 11° and 23° S. J Fish Biol 75:797–815. https://doi.org/10.1111/j.1095-8649.2009.02324.x
Fricke R, Eschmeyer WN, van der Laan R (2022) Eschmeyer’s Catalog of Fishes: Genera, Species, References. Electronic version. http://researcharchive.calacademy.org/research/ichthyology/catalog/fishcatmain.asp. Accessed 11062022
Garman S (1899) The Fishes. Mem Mus Comp Zool 24:1–431
GEBCO (2014) The GEBCO_2014 Grid, version 20150318. In: Gen. Bathymetr. Chart Oceans. http://www.gebco.net. Accessed 2 July 2018
Günther A (1878) Preliminary notices of deep-sea fishes collected during the voyage of H. M. S. "Challenger". Annals and Magazine of Natural History, Series 5, v. 2, nos 7/8/9, art. 2/22/28: 17-28, 179-187, 248-251
Harbour RP, Leitner AB, Ruehlemann C, Vink A, Sweetman AK (2020) Benthic and demersal scavenger biodiversity in the eastern end of the clarion-clipperton zone - an area marked for polymetallic nodule mining. Front Mar Sci 7. https://doi.org/10.3389/fmars.2020.00458
Hein JR, Koschinsky A, Kuhn T (2020) Deep-ocean polymetallic nodules as a resource for critical materials. Nat Rev Earth Environ 1:158–169. https://doi.org/10.1038/s43017-020-0027-0
Holleman W, Fennessy S, Russell B, Maunde C (2020). Ipnops pristibrachium. The IUCN Red List of Threatened Species 2020: e.T123323501A123323740. https://doi.org/10.2305/IUCN.UK.2020-1.RLTS.T123323501A123323740.en. Accessed 11 March 2022
Hubbs CL, Lagler KF (1958) Fishes of the Great Lakes region (revised edn). University of Michigan Press, Michigan 213 pp
International Seabed Authority (2020) Deep seabed mineral resources. https://www.isa.org.jm/exploration-contracts/polymetallic-nodules. Accessed 10 March 2020
International Seabed Authority (2021) Decision of the Council of the International Seabed Authority relating to the review of the environmental management plan for the Clarion-Clipperton Zone. Kingston, Jamaica
Ivanova NV, Zemlak TS, Hanner RH, Hebert PDN (2007) Universal primer coctails for fish DNA barcoding. Molecular Ecology Notes 7:544–548. https://doi.org/10.1111/j.1471-8286.2007.01748.x
Katoh K, Misawa K, Kuma KI, Miyata T (2002) MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res 30:3059–3066. https://doi.org/10.1093/nar/gkf436
Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, Thierer T, Ashton B, Mentjies P, Drummond A (2012) Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28:1647–1649. https://doi.org/10.1093/bioinformatics/bts199
Langenkämper D, Zurowietz M, Schoening T, Nattkemper TW (2017) BIIGLE 2.0 – Browsing and annotating large marine image collections. Front Mar Sci 4:83. https://doi.org/10.3389/fmars.2017.00083
Leitner AB, Durden JM, Smith CR, Klingberg ED, Drazen JC (2021) Synaphobranchid eel swarms on abyssal seamounts: Largest aggregation of fishes ever observed at abyssal depths. Deep Sea Res Part I Oceanogr Res Pap 167:103423. https://doi.org/10.1016/j.dsr.2020.103423
Lewis M (2010) The CSIRO 4 m Beam Trawl. CSIRO Marine and Atmospheric Research Paper, 033, pp 1–17 Available from http://www.cmar.csiro.au/publications/cmarseries/Internal%20Report_BeamTrawl.pdf. Accessed 01 June 2022
Marshall NB, Staiger JC (1975) Aspects of the structure, relationships, and biology of the deep-sea fish Ipnops murrayi (Family Bathypteroidae). Bull Mar Sci 25(I):101–111
McEachran JD, Fechhelm JD (1998) Fishes of the Gulf of Mexico, Vol. 1. Austin, TX: University of Texas Press
Miller KA, Thompson KF, Johnston P, Santillo D (2018) An overview of seabed mining including the current state of development, environmental impacts, and knowledge gaps. Front Mar Sci 4:418. https://doi.org/10.3389/fmars.2017.00418
Moore J, Polanco Fernandez A (2019) Ipnops murrayi. The IUCN Red List of Threatened Species 2019: e.T123323471A123323735. https://doi.org/10.2305/IUCN.UK.2019-2.RLTS.T123323471A123323735.en. Accessed 11 March 2022
Nelson JS, Grande TC, Wilson MVH (2016) Fishes of the World. John Wiley & Sons, Inc., pp 1-752
Nielsen JG (1966) Synopsis of the Ipnopidae (Pisces: Iniomi) with description of two new abyssal species. In Wolff T (ed) Galathea Report 8, Danish Science Press, Ltd., Copenhagen, pp 49–75
Okiyama M, Ida H (2010) Record of Ipnops sp. (Ipnopidae: Aulopiformes) from northern Japan. Ichthyol Res 57:422–423. https://doi.org/10.1007/s10228-010-0171-5
Paxton JR, Niem VH (1999) Ipnopidae. In: Carpenter KE, Niem VH (eds) The Living Marine Resources of the Western Central Pacific. FAO Species Identification Guide for Fishery Purposes Vol. 3, part 1, FAO, Rome, pp 1923–1924
Russell B, Holleman W, Fennessy S, Maunde C (2020) Ipnops meadi. The IUCN Red List of Threatened Species 2020: e.T123323464A123323730. https://doi.org/10.2305/IUCN.UK.2020-1.RLTS.T123323464A123323730.en. Accessed 11 March 2022
Simon-Lledó E, Pomee C, Ahokava A, Drazen JC, Leitner AB, Flynn A, Parianos J, Jones DOB (2020) Multi-scale variations in invertebrate and fish megafauna in the mid-eastern Clarion Clipperton Zone. Prog Oceanogr 187:102405. https://doi.org/10.1016/j.pocean.2020.102405
Sulak KJ (1990) Chlorophthalmidae (Ipnopinae) (pp.353-360), Synodontidae (pp. 365-370). In: Quéro J-C, Hureau J-C, Karrer C, Post A, Saldanha L (1990) Check-list of the fishes of the eastern tropical Atlantic. CLOFETA. UNESCO, Paris, 3 vols. Pp. i-xxxii + 1-1492
Ward RD, Zemlak TS, Innes BH, Last PR, Hebert PDN (2005) DNA barcoding Australia’s fish species. Philos Trans R Soc B 360:1847–1857. https://doi.org/10.1098/rstb.2005.1716
Acknowledgements
Many thanks to Irina Eidus (Leibniz Institute for the Analysis of Biodiversity Change, Hamburg) for her help with the radiographs. The authors would like to thank Carsten Rühlemann (Federal Institute for Geosciences and Natural Resources, BGR, Hanover) for making the material from the BGR cruise BIONOD available. The authors acknowledge the CSIRO Marine National Facility for samples and data utilized in this publication that were collected as part of three RV Investigator voyages and charters in 2015 and 2017. We thank the Chief Investigators, particularly Dr. Alan Williams (CSIRO) and Dr. Timothy O'Hara, (Museum Victoria), the scientific staff, support staff and crew who participated on those surveys. We acknowledge funding support from the Great Australian Bight Research Program (GABRP), a collaboration between BP, CSIRO, the South Australian Research and Development Institute (SARDI), the University of Adelaide, and Flinders University, and the Great Australian Bight Deepwater Marine Program (GABDMP), a CSIRO-led research program sponsored by Chevron Australia, with data generated to be made publicly available. For catch processing and curation of specimens collected during these surveys, we particularly thank Alastair Graham (CSIRO, Hobart), Dianne Bray and Dr. Martin Gomon (Museum Victoria, Melbourne) and Ken Graham (Australian Museum Research Institute, Sydney). We also thank the two anonymous reviewers for providing comments and suggestions that greatly improved the manuscript. This is publication 89 from the Senckenberg am Meer Molecular and Metabarcoding Laboratory.
Funding
Open Access funding enabled and organized by Projekt DEAL.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Ethics approval
All applicable international, national, and/or institutional guidelines for the care and use of animals were followed by the authors.
Sampling and field studies
All necessary permits for sampling have been obtained by the authors from the competent authorities and are mentioned in the Acknowledgements, if applicable. The study is compliant with CBD and Nagoya protocols.
Data availability
The molecular datasets generated and/or analysed during the current study and other publicly available datasets are accessible in the BOLDSYSTEMS repository, www.boldsystems.org. Newly generated and already in BOLD existing nucleotide sequences were also deposited in the GenBank repository, www.ncbi.nlm.nih.gov/genbank, with accession numbers included in this published article.
Author contribution
RT and PMA conceived and designed research. RT and JJP conducted the morphological analyses. MC and SAA carried out the molecular analyses. KU contributed the image annotation data. TW provided photographs of the collected specimen. RT wrote the original draft of the manuscript. AV has critically gone through the manuscript and provided suggestions. All authors read and approved the manuscript.
Additional information
Communicated by T. Horton
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
This article is a contribution to the Topical Collection Biodiversity in Abyssal Polymetallic Nodule Areas
Supplementary information
ESM 1
(DOCX 292 kb)
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Thiel, R., Christodoulou, M., Pogonoski, J.J. et al. An application of morphological analysis and DNA barcoding to identify Ipnops from the Clarion-Clipperton Zone (CCZ) as I. meadi Nielsen, 1966 with notes on other species of the genus (Aulopiformes: Ipnopidae). Mar. Biodivers. 52, 68 (2022). https://doi.org/10.1007/s12526-022-01320-1
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s12526-022-01320-1