Abstract
The global increase of toxin-producing cyanobacteria poses a serious risk to humans. Many investigations have shown that the cyanotoxin microcystin-LR induces hepatotoxicity in rodents. However, many of these studies applied the toxin intraperitoneally or used high oral concentrations, leading to an unrealistically high bioavailability of the toxin. Such approaches have put into question how these results translate to human exposure scenarios. Epidemiology studies have linked microcystin-LR with hepatotoxicity and liver cancer in humans, though by design these investigations cannot provide direct evidence. The present work investigated the effect of microcystin-LR exposure on pigs closely mimicking real-life human conditions. In two animal experiments, pigs were administered microcystin-LR daily by oral gavage for 35 days. Metabolomic and lipidomic tools were used to analyse blood and liver samples. In addition, blood biochemistry parameters indicative of liver function and health were studied to further investigate the potential hepatotoxic effects of microcystin-LR. Results indicated that the metabolomic and lipidomic analyses did not show a gross treatment effect in blood and liver. Furthermore, no significant alterations were found in the tested blood biochemistry parameters. No evidence of hepatotoxicity was found. These results shed more light onto the effects (or lack of effects) of low-dose oral microcystin-LR exposure. The data suggests that the risk of oral microcystin-LR exposure may be overestimated.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The global increase of toxin-producing cyanobacteria in freshwater systems is a growing concern. Also known as blue-green algae, cyanobacteria are photosynthetic prokaryotes present in lakes, ponds and rivers across the world (van Apeldoorn et al. 2007). Some genera produce secondary metabolites known as cyanotoxins that are harmful to the environment and human health. The most widely studied cyanotoxins belong to the microcystin class, of which microcystin-LR (MC-LR) is believed to be the most toxic variant. It binds to the catalytic centre of serine/threonine protein phosphatases 1 and 2A (Runnegar et al. 1995), thereby inhibiting protein phosphatase activity (Hastie et al. 2005; MacKintosh et al. 1990) and perturbing the phosphorylation/dephosphorylation balance in cells. This affects the various cellular functions PP1 and PP2A which are involved in Gehringer (2004). Mechanisms affected by MC-LR exposure include apoptosis (Chen and Xie 2016) and cytoskeletal organization (Zhou et al. 2015). MC-LR has also been found to cause oxidative stress by inducing the formation of reactive oxygen species, leading to genotoxicity (Zegura 2016). Two-stage carcinogenesis experiments also have shown that MC-LR can act as a tumour promoter in vivo (Nishiwaki-Matsushima et al. 1992; Ohta et al. 1994; Sekijima et al. 1999; Zegura 2016), and the International Agency for Research on Cancer (IARC) has classified MC-LR as a group 2B carcinogen, i.e. MC-LR is possibly carcinogenic to humans (IARC Working Group on the Evaluation of Carcinogenic Risks to Humans 2010).
Furthermore, multiple animal toxicology studies in mammals described dose-dependent hepatic injury and altered changes in liver function enzyme levels after oral MC-LR exposure (Fawell et al. 1999; Heinze 1999; Sedan et al. 2015), though toxic effects were not always found (Ueno et al. 1999). In a study performed by Fawell and colleagues, mice were treated with MC-LR by oral gavage for 13 weeks daily using various concentrations (Fawell et al. 1999). The no-observed-adverse-effect level (NOAEL) was 40 µg/kg bodyweight. Based on this NOAEL, the World Health Organization (WHO) applied an uncertainty factor of 1000 to the NOAEL (100 for intra- and interspecies variation, 10 for limitations in the database) and set the tolerable daily intake (TDI) at 0.04 µg/kg bodyweight per day (Chorus and Bartram 1999).
However, several epidemiology studies have reported signs of hepatotoxicity in populations exposed to MC-LR amounts similar to the TDI. Chen and colleagues showed a positive correlation between microcystin serum concentrations in local fishermen from Lake Chaohu and several liver function enzymes (Chen et al. 2009). The authors estimated that the fishermen were exposed to MC-LR equivalent concentrations similar to the MC-LR TDI. Furthermore, Li et al. discovered that children who consumed drinking water sourced from microcystin-contaminated lakes had significantly higher levels of liver function enzymes than the control population (Li et al. 2011). Exposure to microcystins has also been linked to an increased incidence of primary liver cancer (Ueno et al. 1996) and colorectal cancer (Lun et al. 2002).
In toxicology, advancements in -omics studies have allowed the identification and quantification of biomarkers in response to xenobiotics. A large number of studies has looked at the effect of MC-LR on the rodent proteome (Chen et al. 2005; He et al. 2017; Li et al. 2012a, b; Li et al. 2012a, b; Zhao et al. 2012, 2015, 2016), transcriptome (Bulera 2001; Chen et al. 2005; Clark et al. 2007; Zhang et al. 2016) and metabolome (Cantor et al. 2013; He et al. 2012, 2017; Zhang et al. 2016), in which each study reported changes in the treated group as opposed to the control group. However, the lowest concentration used in studies investigating MC-LR toxicity in rodents by oral administration was 40 µg/kg bodyweight (He et al. 2012, 2017; Zhang et al. 2016), which is a thousand times higher than the TDI. Furthermore, many -omics studies applied the toxin by intraperitoneal injection (IP), which is an unrealistic route of exposure in humans. While the results from these studies are relevant for a better understanding of MC-LR’s modes of action, the toxin administration methods used call into question whether the results are meaningful for reliable human risk assessments.
In order to assess the risk of MC-LR exposure in humans, it is necessary to use realistic exposure conditions (i.e. low toxin concentrations dosed by oral administration) and organisms that are as similar to humans as realistically feasible. Here, we present the results of two independent -omics investigations in pigs that were treated with 2.0 (50 × TDI) and 8.0 (200 × TDI) MC-LR µg/kg bodyweight by oral gavage (referred to as animal experiment #1 and #2, respectively). The pig was chosen as the translational model because it is anatomically and physiologically similar to humans (Bassols et al. 2014). Blood samples from animal experiment #2 underwent metabolomic analysis, whereas liver samples from both animal experiments were analysed at the metabolome level. Furthermore, the lipidome of liver samples from animal experiment #2 was investigated as well. Ultra-performance liquid chromatography coupled with high-resolution mass spectrometry (UPLC-HRMS) was used to interrogate the metabolomes and lipidomes. Parallel to the -omics work, blood biochemistry parameters indicative of liver function and health were studied to further investigate the hepatoxic effects of MC-LR. The findings presented here shed more light onto the effects (or lack of effects) of orally dosed MC-LR using concentrations relevant to real-life human exposure conditions.
Materials and Methods
All solvents (LC–MS grade or similar) were purchased from Sigma-Aldrich. Water was supplied from an in-house 18 MΏ Millipore water system (Millipore). Lasalocid A sodium salt solution stock solution (100 µg mL−1 in acetonitrile, analytical standard) was purchased from Sigma-Aldrich (Catalogue Number 33339, Batch Number BCBV9111). Purified MC-LR was purchased from Enzo Life Sciences Ltd.
Animal Experiment #1
The animal experiment was performed as described by Greer et al. (2018). Twelve pigs participated in the study (Duroc breed). Each individual animal was housed in a separate pen that met the requirements of EC/2010/63 Directive regarding animal welfare. The pens were made of chain link and contained sawdust and wood shavings on the floor. This enabled the animals to root and prevented slipping. The pigs had access to chains that they could play with. The animals were placed in sight of each other.
The diet was based on breeder’s recommendations. The animals were fed the same quantity of food each day. The amount was determined by the weight of the control group, and the food intake was recorded daily. The animals had free access to water, which was provided by an automated watering system.
The animals were acclimatized for two weeks, after which six pigs were randomly selected for the control group. The remaining six pigs were assigned to the treatment group. A veterinarian examined the pigs before weighing them. The weight and identification of each animal was recorded. For 35 days, each animal from the treatment group was dosed with MC-LR by oral gavage (2.0 µg/kg bodyweight, or 50 × TDI). A 1 mg/mL solution of MC-LR was prepared by the addition of 250 μL ethanol followed by 750 μL water. The weight of each animal was subsequently used to prepare the appropriate dilutions. The dose was administered in 5 mL water.
At the end of the experiment, all animals were euthanized by intravenous barbiturate and the pigs exsanguinated by jugular severance. Liver samples were collected and stored at − 80 ºC.
Complications arose during the animal study. One animal from the control group and one pig from the treated group became ill and were treated with appropriate drugs. Because these animals received additional treatments, they were excluded from the -omics analyses.
Animal Experiment #2
This study was performed as described for animal experiment #1 using sixteen male pigs (a cross of Large White and Landrace breeds, PIC 337 breed), eight control and eight treated pigs. Housing, diet and treatment were as described previously with modifications.
At the beginning of the study, control pig #6 developed a hernia and was put down as directed by the veterinarian. A substitute control pig was introduced before treatment commenced.
Over the study period, each animal from the treatment group received a daily dose of 8 µg toxin/kg body weight MC-LR by oral gavage, 200 times the provisional TDI set by the WHO. MC-LR dosing was prepared as described above. Blood was collected weekly using a Vacutainer system. Care was taken to process the samples as quickly as possible, after which they were stored at − 80 ºC. The last blood collection was carried out on day 35. At the end of the experiment, the animals were euthanized and liver samples were collected as outlined for animal experiment #1.
Blood Metabolomics: Metabolite Extraction
The plasma extraction protocol was based on previously published methods (Arias et al. 2016; Graham et al. 2013) with minor modifications. In order to extract metabolites from the plasma samples (animal experiment #2), 400 µL of ice-cold methanol was added to 100 µL of blood sample in random order. After rigorous vortexing for 10 min, the samples were left to chill on ice for 10 min, followed by centrifugation at 15,000g for 10 min at 4 ºC. The supernatant was collected and evaporated to dryness at 30 ºC using a centrifugal vacuum concentrator (miVac Quatro Concentrator, Mason Technology, Dublin, Ireland).
Liver Metabolomics: Metabolite Extraction
As with the plasma samples, the extraction protocol was adapted from previously published methods (Arias et al. 2016; Graham et al. 2013). The liver samples from animal experiment #1 and #2 were lyophilized using a Christ freeze drier (Osterode, Germany). Dried tissue samples from pig experiment #1 were milled under liquid nitrogen using a mortar and pestle, whereas the liver samples from pig experiment #2 were homogenized under liquid nitrogen using a 6850 Freezer/Mill (SPEX SamplePrep LLC, US) (Meneely et al. 2016). The resulting powdered samples were stored at − 80 ºC until further analysis.
Metabolites were extracted from the liver samples by adding 0.5 mL ice-cold methanol:water (4:1) to 25 mg of liver tissue from animal experiment #1 in random order, whereas for animal experiment #2 1.0 mL of ice-cold methanol:water (4:1) was added to 50 mg of liver tissue in random order. After vortexing, samples were sonicated in an ice-water bath for 15 min (Ultrasonic cleaner USC 600 TH, VWR International, UK) (Arias et al. 2016). This was to preserve the integrity of the metabolites, to increase protein precipitation and to minimize solvent evaporation during ultrasound agitation that led to an increase of water bath temperature. This was followed by centrifugation at 16,000g for 20 min. at 4 ºC, after which the metabolite-containing supernatant was collected and evaporated to dryness in a centrifugal vacuum concentrator at 30 °C (miVac Quatro Concentrator, Mason Technology, Dublin, Ireland). Extracts were kept at − 80 °C until UPLC-HRMS analysis.
Liver Lipidomics: Lipid Extraction
The lipid extraction protocol was based on a previously published protocol (Chen et al. 2013), with modifications. Briefly, lipids were extracted from liver samples (animal experiment #2 only) by adding 1 mL ice-cold methyl-tert-butylether:methanol (3:1) to 50 mg of lyophilized liver tissue in random order. The samples were vortexed rigorously for 20 s, followed by 5 min. of sonication in an ice-water bath, after which they were shaken at room temperature for 1 h. To induce phase separation, 450 µL of water was added. The samples were then vortexed briefly and incubated at − 20 ºC for 10 min, followed by centrifugation at 14,000g for 15 min at 4 ºC. The lipid-containing upper phase was collected and evaporated to dryness in a centrifugal vacuum concentrator at 30 °C (miVac Quatro Concentrator, Mason Technology, Dublin, Ireland). Extracts were kept at − 80 °C until UPLC-HRMS analysis.
UPLC-HRMS Analysis
The metabolomics samples were reconstituted in water and filtered by centrifugation using 0.22 µm Constar Spin-X centrifuge tube filters (Corning Incorporated). The lipidomics samples (liver, animal experiment #2 only) were reconstituted in methanol and filtered using Whatman Mini-UniPrep G2 filters (GE Healthcare). For each UPLC-HRMS analysis, a pooled sample was prepared by combining equal volumes of each sample. Tables 1 and 2 list the details of each UPLC-QToF and UPLC-Orbitrap analysis per sample type and animal experiment, respectively.
The column was equilibrated for each UPLC-HRMS analysis by injecting at least ten pooled samples at the start. For quality control, pooled samples were injected at intervals of every five samples throughout the entire experiment to determine the chromatographic reproducibility and peak intensities. Each sample was injected three times at random in order to confirm technical reproducibility (Graham et al. 2013).
Data Processing and Multivariate Statistical Analysis
All raw data were processed using Progenesis QI software (version 2.3, Waters, UK). The software selected the most suitable injection of the pool sample as alignment reference. Peaks were then automatically aligned and manually reviewed, and ion intensities were normalized against all compounds. Each run was filtered by performing noise reduction (in the form of a data import filter) and by filtering at the peak picking stage. The filter settings applied for each UPLC-HRMS analysis are outlined in Tables 1 and 2. The normalized data from all three replicate and pool injections were exported from Progenesis QI and imported into Simca 15.0 (Umetrics, Umea, Sweden) to assess the stability of the UPLC-HRMS instrument. PCA was applied to the data to verify that the pooled samples clustered at the centre of the scores plot, giving a good indication of data stability and reproducibility (Graham et al. 2013). The grouping of technical replicates was verified as well. For further analysis, the raw data of one set were re-processed in Progenesis QI using the settings listed in Tables 1 and 2 for each UPLC-HRMS analysis. Peak picking was disabled for the pooled samples. The data were then exported to Simca for multivariate analysis using unsupervised PCA and supervised OPLS-DA models. These models generate an R2 (an estimation of the model’s fit) and Q2 value (an estimation of the model’s predictive capabilities), which are automatically calculated by cross-validation in Simca. The R2 and Q2 cum values were used to determine the validity of the model. Within the Simca software environment, significance of each OPLS-DA model was tested using CV-ANOVA.
Univariate Statistical Analysis
Within the Progenesis QI software environment, a one-way analysis of variance (ANOVA) test was carried out to assess significant increases or decreases of ion abundancies when comparing the control and treated groups. An FDR adjusted p-value (or q-value) of 0.01 was chosen to minimize the likelihood of finding false positives, as well as an absolute fold-change cut-off of 2. Each peak was manually reviewed in the raw data to ascertain it was genuine. Where available, MS2 spectra were queried to the mzCloud database (https://www.mzcloud.org/) to obtain putative identifications.
Identification of Lasalocid A by UPLC-MS/MS Analysis
Lasalocid A was extracted from the liver samples from animal experiment #2 using the lipid extraction strategy as outlined previously.
Analysis of lasalocid A was performed using an ACQUITY UPLC i-Class FTN system coupled to a Xevo Triple-Quadrupole-MS/MS (Waters, Manchester, UK). The system was operated in both ESI+ and ESI− modes with a capillary voltage of 1.5 kV for both. Source and desolvation temperatures were 150 and 400 °C, respectively, with the desolvation gas flow set at 700 L/h. Detection was achieved using targeted analysis via multiple reaction monitoring (MRM) involving fragmentation of specific precursor ions (parent) using argon as the collision gas, to at least two product ions (daughters), with the cone voltage and collision energies optimized manually, indicated in Table S1.
Separation was achieved using an ACQUITY UPLC BEH C18 column, 50 mm × 2.1 mm i.d., 1.8 µm particle size, 130 Å pore size, (Waters, UK) maintained at 40 °C. The mobile phases consisted of water and methanol both containing 0.1% formic acid (v/v). The flow rate was set at 0.4 mL/min with the methanol held at 15% for 0.5 min, followed by an increase to 40% over 2.5 min. The methanol content was increased further to 95% over the next 1.5 min, maintained for 2.5 min before returning to 15% for a 1 min re-equilibration before the next injection. The injection volume was set at 2.5 µL.
Blood Biochemistry Parameters
For animal experiment #2, levels of AST, ALP, ALT, GGT, LDH, albumin and total protein were measured in plasma samples from day 35. This was carried out by the Centre for Public Health at Queen’s University Belfast (UK) using an RX Daytona + clinical chemistry analyzer (Randox Laboratories, UK). A two-tailed, two-sample, equal-variance Student’s t-test was carried out to search for significant concentration differences between the control and treated animals.
Results
Blood Metabolomics: Multivariate Statistical Analysis
The plasma samples from animal experiment #2 were investigated at the metabolome level using a UPLC-Quadrupole Time of Flight mass spectrometer (QToF). Table 1 lists the experimental details. Multivariate models were constructed to analyse differences between the control and treated groups. No effects were witnessed (Fig. 1). Visually, the unsupervised principal component analysis (PCA) model indicated that the within-group variance was larger than the between-group variance (Fig. 1a), since no distinct grouping was witnessed. The associated Q2 cumulative (cum) values were low, indicating weak model predictability. In addition, a supervised orthogonal projection to latent structures discriminant analysis (OPLS-DA) model was built to assess the data in more detail (Fig. 1b). Significance of the OPLS-DA model was tested using an ANOVA of the cross-validated residuals (CV-ANOVA). Indeed, while the model visually showed separation and a higher Q2 value, the results of the CV-ANOVA test indicated that the model was not statistically significant (Fig. 1c). The results from earlier treatment days were similar to those obtained for day 35 and can be found in the supplementary data file (Figs. S1–S3). At the start of animal experiment #2, one pig from the control group developed a hernia and therefore its blood sample was excluded from the metabolomic analysis of the pre-bleed sample (Fig. S1).
Liver Metabolomics: Multivariate Statistical Analysis
The metabolome of the liver samples from animal experiment #1 and #2 were analysed using a UPLC-QToF (Table 1). One sample from animal experiment #1 (control group) was excluded due to limited sample availability. Similar to the results found for the blood metabolome, multivariate analysis of the liver metabolomes yielded PCA models with larger within-group variance than between-group variance, as indicated by the fact that the data points did not form distinct groups (Fig. 2a, b). While for animal experiment #1 the associated Q2(cum) value indicated a degree of model predictability (Q2(cum) = 0.337), an OPLS-DA model showed poor model predictability (Q2(cum) = -0.394, Fig. 2c). The OPLS-DA model for the animal experiment #2 data also indicated weak model predictability (Fig. 2d). Subsequent CV-ANOVAs of the OPLS-DA models indicated no statistical significance in both datasets (Fig. 2e, f).
Multivariate analysis of UPLC-QToF liver metabolomics data from animal experiment #1 (ESI+) and animal experiment #2 (ESI+). PCA models of experiment #1 (a) and experiment #2 samples (b). OPLS-DA models of experiment #1 (c) and experiment #2 samples (d), CV-ANOVAs of the OPLS-DA models for experiment #1 (e) and experiment #2 (f). Blue dots, control. Violet dots, treated
The metabolomes of the liver samples from animal experiment #2 were also analysed using a UPLC-Orbitrap in both ESI+ and ESI− modes (Table 2). In both datasets, the within-group variance was larger than the between-group variance on the PCA plots (Fig. 3a, b). Furthermore, the OPLS-DA models and CV-ANOVAs indicated that the models had poor predictive capabilities and were statistically non-significant (Fig. 3c–f).
Multivariate analysis of UPLC-Orbitrap liver metabolomics data from animal experiment #2 (ESI+ and ESI−). PCA models of ESI+ dataset (a) and ESI− dataset (b), OPLS-DA models of ESI+ dataset (c) and ESI− dataset (d), CV-ANOVAs of the OPLS-DA models for ESI+ (e) and ESI− (f). Blue dots, control. Violet dots, treated
Liver Lipidomics: Multivariate Statistical Analysis
The liver samples from animal experiment #2 were analysed at the lipidome level using a UPLC-QToF and a UPLC-Orbitrap using both ESI+ and ESI− modes (Tables 1 and 2). The results obtained from the multivariate analyses were similar to those from the liver metabolome. For the QToF data, PCA plots built from both the ESI+ and ESI− datasets visually showed no distinct grouping (Fig. 4a, b). Subsequent OPLS-DA models (Fig. 4c, d) and CV-ANOVAs (Fig. 4e, f) indicated that the models had low predictive capabilities and were statistically non-significant. The data derived from the Orbitrap analysis were similar to the QToF data (Fig. 5a–f): none of the multivariate models indicated a treatment effect.
Multivariate analysis of UPLC-QToF liver lipidomics data from animal experiment #2 (ESI+ and ESI−). PCA models of ESI+ dataset (a) and ESI− dataset (b), OPLS-DA models of ESI+ dataset (c) and ESI− dataset (d), CV-ANOVAs of the OPLS-DA models for ESI+ (e) and ESI− (f). Blue dots, control. Violet dots, treated
Multivariate analysis of UPLC-Orbitrap liver lipidomics data from animal experiment #2 (ESI+ and ESI−). PCA models of ESI+ dataset (a) and ESI− dataset (b), OPLS-DA models of ESI+ dataset (c) and ESI− dataset (d), CV-ANOVAs of the OPLS-DA models for ESI+ (e) and ESI− (f). Blue dots, control. Violet dots, treated
Univariate Statistical Analysis
In addition to the multivariate statistics, univariate statistical analyses were carried out. Within the Progenesis QI software environment (version 2.3, Waters, UK), a one-way ANOVA test with false discovery rate (FDR) correction was performed on each dataset to assess significant increases or decreases of ion abundancies when comparing the control and treated groups (q < 0.01; fold-change > 2). The MS2 spectra of these features were submitted the mzCloud database (https://www.mzcloud.org/), but no reliable putative identifications were made for most compounds (level 4 according to the minimum reporting standards for chemical analysis guidelines (Sumner et al. 2007)). However, one feature was putatively identified as lasalocid A (Table S2). The ANOVA details for this feature are listed in Table S3. Using a lasalocid sodium standard, the identity of lasalocid A was verified by UPLC-Triple-Quadrupole-MS at level 1 according to the minimum reporting standards for chemical analysis guidelines (Sumner et al. 2007). Samples were screened for the following lasalocid A adducts: M + H, M − H and M + Na + (Table S1). While M + H was not detected in any of the tested samples, the presence of M − H and M + Na + were verified in the treated samples (Fig. S4a, treated animal #5, M + Na + shown). In line with the lipidomics results, the tested control samples only contained trace amounts of lasalocid A (Fig. S4b, control animal #5, M + Na + shown).
Blood Biochemistry Parameters
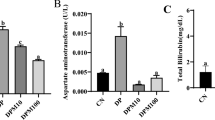
For animal experiment #2, levels of aspartate aminotransferase (AST), alkaline phosphatase (ALP), alanine transaminase (ALT), gamma-glutamyl transferase (GGT), lactate dehydrogenase (LDH), total protein and albumin were measured for the day 35 (last treatment day) plasma samples (Table 3). A two-tailed, two-sample, equal-variance Student’s t-test did not detect a significant difference between the control and treated groups for any of the measured parameters (p < 0.05, see Table S4).
Discussion
This study mimicked real-life human MC-LR exposure conditions by treating pigs orally with low toxin concentrations. The pig was chosen as the animal model because of its anatomical and physiological similarity to humans (Bassols et al. 2014). Based on the -omics analyses presented here, there is no evidence that the MC-LR concentrations administered orally have a toxic effect on the blood metabolome, and neither on the liver metabolome and lipidome.
Multivariate Statistical Analysis of All -Omics Data
Blood samples from animal experiment #2 (200 × TDI), as well as liver samples from animal experiment #1 (50 × TDI) and #2 were analysed at the metabolome level. Liver samples from animal experiment #2 also underwent lipidomic analysis. In order to analyse the vast amount of ion abundancy data generated by such experiments, multivariate statistics are often used. Generally, an unsupervised PCA model is considered the first “checkpoint” a dataset must pass in order to continue downstream analysis (Worley and Powers 2016). If distinct clustering of groups is witnessed on a PCA plot, this suggests a treatment effect. On the other hand, scrambled data points and no grouping are indicative of there not being a (significant) experimental effect. None of the PCA models generated in this study showed any distinct grouping of the control and treated groups. The associated Q2(cum) values of most of the models were low, indicating weak predictive capabilities.
Even though the PCA models showed that there was no treatment effect, supervised OPLS-DA models were built to further study the data. OPLS-DA plots have been shown previously to cluster data points into groups using random data (Kjeldahl and Bro 2010; Worley and Powers 2013), and visually they are not reliable to infer conclusions on biological effects from. As expected, all models illustrated clustering of the control and treatment groups, but again in most cases the Q2(cum) values were close to 0 or even negative. CV-ANOVA tests were performed to check whether the OPLS-DA models were statistically significant, but this was not the case for any model, and in nearly all cases the p-value was (close to) 1. Together, the results from the multivariate statistics generated by both the PCA and OPLS-DA models show that relatively low doses of oral MC-LR exposure have no (significant) effect on the gross blood and liver metabolome of pigs, and neither on the liver lipidome.
These results contradict the current literature, where MC-LR has been reported to cause an effect in the blood metabolome (He et al. 2017; Zhang et al. 2016). Zhang and colleagues treated mice with MC-LR by oral gavage for 28 days, using concentrations of 40 and 200 µg/kg bodyweight, i.e. 1000× and 5000× TDI, respectively (Zhang et al. 2016). They built PLS-DA models using the obtained spectra and found separation between control and treated mice, identifying several serum metabolites whose abundancies were altered (Zhang et al. 2016). In another study, He et al. treated mice using the same concentrations as Zhang and colleagues, but applied the toxin every 2 days for 90 days by oral gavage (He et al. 2017). The OPLS-DA plots presented in their work show separation between control and treated groups, as well as Q2(cum) values of 0.557 and higher (He et al. 2017). Indeed, both studies found a treatment effect, contrary to the results presented here.
Experiments on rodents have also been used to analyse the effect of MC-LR toxicity on the liver metabolome (He et al. 2012, 2017). Similar to the blood metabolomics studies cited previously, the reports describe very high toxin doses. Indeed, the lowest oral dose administered was 40 µg/kg bodyweight (He et al. 2012, 2017). This equates to being a thousand times higher than the TDI set by the WHO. While these studies are useful for gaining further insights into MC-LR’s mode of action, they are arguably less suitable for reliable human risk assessment.
Because the present investigation aimed to mimic human exposure conditions as closely as possible, meaning low MC-LR concentrations were administered orally, the bioavailability of the toxin may have been too low to exert a significant effect on any of the investigated -omes. However, there is another possible explanation for the discrepancy between the literature and this study. Up until now, all -omics investigations used rodents as the mammalian model to analyse MC-LR toxicity, whereas this study describes the effect of MC-LR on pigs. Rodents and pigs are very different from each other, with pigs being anatomically and physiologically much more similar to humans (Bassols et al. 2014). The MC-LR uptake mechanisms and subsequent toxic effects may be different for rodents than they are for mammals more closely resembling humans. In fact, a study by Fawell and colleagues found that mice were more sensitive to MC-LR than rats (Fawell et al. 1999), which indicates that even closely related species may have different degrees of vulnerability to the same toxin.
Univariate Statistical Analysis of All -Omics Data
When analysing ion abundancy data from metabolomics and lipidomics experiments, often OPLS-DA plots are generated that allow the construction of S-plots, which reveal the most discriminatory features that define the OPLS-DA model. The OPLS-DA models generated in this study were of insufficient quality and could therefore not be used to find discriminatory ions. Alternatively, within the Progenesis QI software environment (version 2.3, Waters, UK), univariate statistical analysis of each dataset was performed. Because each dataset contained at least a thousand features, a p-value cut-off did not seem suitable. For example, if a dataset contained 4000 features and a p-value of 0.01 was chosen, then theoretically 1% of 4000 features (40) would have been false positives. Therefore, an FDR adjusted p-value (also known as q-value) of 0.01 was chosen to decrease the likelihood of finding false positives, as well as an absolute fold-change cut-off of 2.
One of the features that showed significant abundancy changes was identified as lasalocid A, which is a coccidiostat commonly used as antibiotic in poultry feed. To our knowledge, it is not used for porcine feed, meaning lasalocid A may have ended up in the pig feed used in this experiment through cross contamination with a feed used for a different animal species. Nevertheless, the result is interesting because it begs the question why the treated animals were not able to clear lasalocid A, at least to the same extent, from their livers, as opposed to the control animals where lasalocid A was present in only trace amounts. Lasalocid A has been shown to perturb sodium fluorescein intake by inhibiting organic anion transporters (OATP) 1B1 and 1B3 in Chinese hamster ovary cells transfected with OATP1B1 and OATP1B3 (De Bruyn et al. 2013). These transporters are involved in the transport of bile salts (Hagenbuch and Meier 2003), but they are also responsible for MC-LR uptake into hepatocytes (Fischer et al. 2005; Komatsu et al. 2007). Perhaps an interaction between MC-LR and lasalocid A may have led to the bioaccumulation of lasalocid A in liver.
Blood Biochemistry Parameters
This investigation measured the levels of blood biochemistry parameters that are typically used to assess liver damage (Table 3). In line with the -omics data presented in this report, there was no significant concentration difference between the control and treated groups for any of the measured parameters (p > 0.05, Table S4). Together with the -omics data, these results suggest that daily oral exposure to 8.0 µg MC-LR/kg bodyweight for 35 days does not adversely affect liver health. These results contradict reports from various toxicology studies that did find significantly altered blood biochemistry parameters after oral exposure to MC-LR (Fawell et al. 1999; Heinze 1999), however, these investigations applied higher toxin concentrations and used rodents.
How Harmful is MC-LR Exposure?
The pigs used in this study did not show signs of liver damage after oral exposure to low MC-LR concentrations. As discussed previously, the discrepancy between this investigation and the published -omics reports describing MC-LR toxicity may be due to the more realistic toxin concentrations used, as well as the translational model being closer to humans.
A study by Falconer and colleagues analysed the effect of oral administration of Microcystis aeruginosa bloom material in drinking water of growing pigs (Falconer et al. 1994). Over a period of 44 days, four groups each consisting of 5 pigs received different concentrations of a Microcystis aeruginosa extract collected from Lake Mokoan, Victoria, Australia. Per dosage group, the microcystin concentrations consumed by the pigs were estimated to be 0 (control group), 280, 796 and 1312 µg/kg bodyweight daily. It is important to note that high-performance liquid chromatography separation revealed the presence of seven microcystin congeners, with microcystin-YR being tentatively identified as the major constituent, though the presence of MC-LR was not detected. Biochemical analysis of the plasma samples revealed that liver function parameters ALT, AST, LDH and total protein were altered in the two highest dosage groups, but no changes were found in the lowest dosage group (280 µg/kg bodyweight). Histopathological examination of the livers revealed that when compared to the control group, most pigs of the two highest treatment groups had signs of hepatic injury, compared to only one pig in the lowest treatment group. The lowest-observed-adverse-effect level (LOAEL) was therefore established at 280 µg/kg bodyweight. While the results are in line with the -omics results presented in the current paper, where no gross effects have been found in blood and liver at a concentration of 8 µg MC-LR/kg bodyweight, it is important to point out that MC-LR was not detected in the bloom extract used by Falconer and colleagues (Falconer et al. 1994). This makes it difficult to compare their results with the current study.
Importantly, low concentrations of orally dosed MC-LR not inducing toxic effects in mammals is not a completely new finding. In 1999, Ueno and colleagues published a report describing no toxicity was witnessed in mice chronically exposed to low MC-LR amounts (20 µg/L) in their drinking water (Ueno et al. 1999). Serum biochemistry parameters and histopathology were analysed, but no significant effects were found after 3, 6, 12 and 18 months of daily exposure (Ueno et al. 1999). More recently, a study published by Labine and colleagues reported that mice exposed to MC-LR in drinking water (1.0 µg/L) for 28 weeks did not reveal signs of hepatotoxicity after examination of liver biochemical parameters and histology (Labine et al. 2017). The results from these investigations are in line with the findings of the present study, where no detrimental effects were witnessed in blood biochemistry parameters and investigated -omes after low-dose oral MC-LR exposure.
It is important to note that the MC-LR concentrations described in this report are far below the reported NOAEL of 40 µg/kg bodyweight (Fawell et al. 1999). This may be why no effects were witnessed. However, the animal experiment was purposefully designed to mimic real-life human exposure conditions, and the concentrations described in this report are more applicable to real-life human scenarios than the NOAEL. However, the results also reveal that the TDI derived from the NOAEL may be appropriate, since no toxic effects were witnessed at concentrations below the NOAEL.
Various epidemiology studies have been published that report a correlation between elevated liver function enzyme levels and frequent oral exposure to water and products from waterbodies contaminated with MC-LR (Chen et al. 2009; Li et al. 2011). MC-LR equivalents were detected in the blood of most subjects exposed to contaminated waters and/or foods (Chen et al. 2009; Li et al. 2011). While these are interesting and indeed worrying findings, they do not provide evidence that MC-LR is the (sole) contributor to the measured changes in liver function enzyme levels. Microcystins alone comprise of more than 100 congeners (Niedermeyer 2014), and these contaminated waters are most likely home to many different groups of (cyanobacterial) toxins. This means that while the correlation between increased MC-LR exposure and altered liver function enzyme levels exists, the studies do not provide proof that MC-LR is a contributing factor of this correlation. The effects witnessed may be a result of the participants being exposed to a cocktail of (cyanobacterial) toxins.
The results presented in this investigation suggest that MC-LR, by itself and at low concentrations, is not toxic to mammals closely resembling humans in terms of liver function. However, this does not mean that the TDI currently set by the WHO (0.04 µg/kg bodyweight) is inappropriate. The TDI is intended to be protective, and since no adverse effects were found in this study, the TDI appears to fulfil its protecting role well. It should also be pointed out that while these experiments suggest that low MC-LR concentrations are not hepatotoxic, in real life humans are exposed to toxin cocktails. While it is not its official function, the MC-LR TDI may serve as a useful indicator that helps estimate the presence of other microcystin congeners or other mixtures of toxins. Because indeed, if MC-LR is detected in a water body, then it is likely that other microcystin congeners are present as well. Consuming cocktails of microcystin congeners, even at low concentrations, may very well induce toxic effects. To better understand this problem, it would be important to study the effects of low-dose administration of MC-LR equivalents in animals similar to humans. This would better mimic real-life human exposure conditions, because humans are exposed to a cocktail of (cyanobacterial) toxins when consuming water and products derived from contaminated water bodies.
Conclusions
The present study revealed that oral exposure to low concentrations of MC-LR (2.0 µg MC-LR/kg bodyweight for 35 days and 8.0 µg MC-LR/kg bodyweight for 35 days) did not induce signs of hepatotoxicity in pigs. Multivariate analyses of the metabolomes and lipidomes of blood and liver samples did not show significant differences between the control and treated animal groups. In addition, blood biochemistry markers indicative of liver health were not significantly affected in the treated animals when compared to the control group. These results shed more light onto the effect of low-dose MC-LR exposure in the context of real-life human conditions and suggest that the risk of oral microcystin-LR exposure may be overestimated.
References
Arias M, Chevallier OP, Graham SF, Gasull-Gimenez A, Fodey T, Cooper KM, Crooks SRH, Danaher M, Elliott CT (2016) Metabolomics reveals novel biomarkers of illegal 5-nitromimidazole treatment in pigs. Further evidence of drug toxicity uncovered. Food Chem 199:876–884. https://doi.org/10.1016/j.foodchem.2015.12.075
Bassols A, Costa C, Eckersall P, Osada J, Sabrià J, Tibau J (2014) The pig as an animal model for human pathologies: a proteomics perspective. Proteomics Clin Appl 8:715–731. https://doi.org/10.1002/prca.201300099
Bulera S (2001) RNA expression in the early characterization of hepatotoxicants in Wistar rats by high-density DNA microarrays. Hepatology 33:1239–1258. https://doi.org/10.1053/jhep.2001.23560
Cantor GH, Beckonert O, Bollard ME, Keun HC, Ebbels TMD, Antti H, Wijsman JA, Bible RH, Breau AP, Cockerell GL, Holmes E, Lindon JC, Nicholson JK (2013) Integrated histopathological and urinary metabonomic investigation of the pathogenesis of microcystin-LR toxicosis. Vet Pathol 50:159–171. https://doi.org/10.1177/0300985812443839
Chen L, Xie P (2016) Mechanisms of microcystin-induced cytotoxicity and apoptosis. Mini-Rev Med Chem 16:1018–1031
Chen T, Wang Q, Cui J, Yang W, Shi Q, Hua Z, Ji J, Shen P (2005) Induction of apoptosis in mouse liver by microcystin-LR a combined transcriptomic, proteomic, and simulation strategy. Mol Cell Proteom 4:958–974. https://doi.org/10.1074/mcp.M400185-MCP200
Chen J, Xie P, Li L, Xu J (2009) First identification of the hepatotoxic microcystins in the serum of a chronically exposed human population together with indication of hepatocellular damage. Toxicol Sci 108:81–89. https://doi.org/10.1093/toxsci/kfp009
Chen S, Hoene M, Li J, Li Y, Zhao X, Häring H-U, Schleicher ED, Weigert C, Xu G, Lehmann R (2013) Simultaneous extraction of metabolome and lipidome with methyl tert-butyl ether from a single small tissue sample for ultra-high performance liquid chromatography/mass spectrometry. J Chromatogr A 1298:9–16. https://doi.org/10.1016/j.chroma.2013.05.019
Chorus I, Bartram J (1999) Toxic cyanobacteria in water: a guide to their public health consequences, monitoring, and management. E & FN Spon, London
Clark SP, Davis MA, Ryan TP, Searfoss GH, Hooser SB (2007) Hepatic gene expression changes in mice associated with prolonged sublethal microcystin exposure. Toxicol Pathol 35:594–605. https://doi.org/10.1080/01926230701383210
De Bruyn T, van Westen GJP, Ijzerman AP, Stieger B, de Witte P, Augustijns PF, Annaert PP (2013) Structure-based identification of OATP1B1/3 inhibitors. Mol Pharmacol 83:1257–1267. https://doi.org/10.1124/mol.112.084152
Falconer IR, Burch MD, Steffensen DA, Choice M, Coverdale OR (1994) Toxicity of the blue-green alga (cyanobacterium) microcystis aeruginosa in drinking water to growing pigs, as an animal model for human injury and risk assessment. Environ Toxic Water 9:131–139. https://doi.org/10.1002/tox.2530090209
Fawell JK, Mitchell RE, Everett DJ, Hill RE (1999) The toxicity of cyanobacterial toxins in the mouse: I microcystin-LR. Hum Exp Toxicol 18:162–167. https://doi.org/10.1177/096032719901800305
Fischer WJ, Altheimer S, Cattori V, Meier PJ, Dietrich DR, Hagenbuch B (2005) Organic anion transporting polypeptides expressed in liver and brain mediate uptake of microcystin. Toxicol Appl Pharm 203:257–263. https://doi.org/10.1016/j.taap.2004.08.012
Gehringer MM (2004) Microcystin-LR and okadaic acid-induced cellular effects: a dualistic response. FEBS Lett 557:1–8. https://doi.org/10.1016/S0014-5793(03)01447-9
Graham SF, Chevallier OP, Roberts D, Hölscher C, Elliott CT, Green BD (2013) Investigation of the human brain metabolome to identify potential markers for early diagnosis and therapeutic targets of Alzheimer’s disease. Anal Chem 85:1803–1811. https://doi.org/10.1021/ac303163f
Greer B, Meneely JP, Elliott CT (2018) Uptake and accumulation of Microcystin-LR based on exposure through drinking water: an animal model assessing the human health risk. Sci Rep 8:4913. https://doi.org/10.1038/s41598-018-23312-7
Hagenbuch B, Meier PJ (2003) The superfamily of organic anion transporting polypeptides. BBA-Biomembranes 1609:1–18. https://doi.org/10.1016/S0005-2736(02)00633-8
Hastie CJ, Borthwick EB, Morrison LF, Codd GA, Cohen PTW (2005) Inhibition of several protein phosphatases by a non-covalently interacting microcystin and a novel cyanobacterial peptide, nostocyclin. BBA-Gen Subjects 1726:187–193. https://doi.org/10.1016/j.bbagen.2005.06.005
He J, Chen J, Wu L, Li G, Xie P (2012) Metabolic response to oral microcystin-LR exposure in the rat by NMR-based metabonomic study. J Proteome Res 11:5934–5946. https://doi.org/10.1021/pr300685g
He J, Li G, Chen J, Lin J, Zeng C, Chen J, Deng J, Xie P (2017) Prolonged exposure to low-dose microcystin induces nonalcoholic steatohepatitis in mice: a systems toxicology study. Arch Toxicol 91:465–480. https://doi.org/10.1007/s00204-016-1681-3
Heinze R (1999) Toxicity of the cyanobacterial toxin microcystin-LR to rats after 28 days intake with the drinking water. Environ Toxicol 14:57–60. https://doi.org/10.1002/(SICI)1522-7278(199902)14:1%3c57:AID-TOX9%3e3.0.CO;2-J
IARC Working Group on the Evaluation of Carcinogenic Risks to Humans (2010) IARC monographs on the evaluation of carcinogenic risks to humans. Ingested nitrate and nitrite, and cyanobacterial peptide toxins. IARC Monogr Eval Carcinog Risks Hum 94(v–vii):1–412
Kjeldahl K, Bro R (2010) Some common misunderstandings in chemometrics. J Chemometr 24:558–564. https://doi.org/10.1002/cem.1346
Komatsu M, Furukawa T, Ikeda R, Takumi S, Nong Q, Aoyama K, Akiyama S, Keppler D, Takeuchi T (2007) Involvement of mitogen-activated protein kinase signalling pathways in microcystin-LR–induced apoptosis after its selective uptake mediated by OATP1B1 and OATP1B3. Toxicol Sci 97:407–416. https://doi.org/10.1093/toxsci/kfm054
Labine M, Gong Y, Minuk GY (2017) Long-term, low-dose exposure to microcystin-LR does not cause or increase the severity of liver disease in rodents. Ann Hepatol 16:959–965. https://doi.org/10.5604/01.3001.0010.5288
Li Y, Chen J, Zhao Q, Pu C, Qiu Z, Zhang R, Shu W (2011) A cross-sectional investigation of chronic exposure to microcystin in relationship to childhood liver damage in the Three Gorges Reservoir Region, China. Environ Health Perspect 119:1483–1488. https://doi.org/10.1289/ehp.1002412
Li G, Cai F, Yan W, Li C, Wang J (2012a) A proteomic analysis of MCLR-induced neurotoxicity: implications for Alzheimer’s disease. Toxicol Sci 127:485–495. https://doi.org/10.1093/toxsci/kfs114
Li G, Yan W, Qiao Q, Chen J, Cai F, He Y, Zhang X (2012b) Global effects of subchronic treatment of microcystin-LR on rat splenetic protein levels. J Proteomics 77:383–393. https://doi.org/10.1016/j.jprot.2012.09.012
Lun Z, Hai Y, Kun C (2002) Relationship between microcystin in drinking water and colorectal cancer. Biomed Environ Sci 15:166–171
MacKintosh C, Beattie KA, Klumpp S, Cohen P, Codd GA (1990) Cyanobacterial microcystin-LR is a potent and specific inhibitor of protein phosphatases 1 and 2A from both mammals and higher plants. FEBS Lett 264:187–192. https://doi.org/10.1016/0014-5793(90)80245-E
Meneely JP, Chevallier OP, Graham S, Greer B, Green BD, Elliott CT (2016) β-methylamino-L-alanine (BMAA) is not found in the brains of patients with confirmed Alzheimer’s disease. Sci Rep 6:36363. https://doi.org/10.1038/srep36363
Niedermeyer T (2014) Microcystin congeners described in the literature. Figshare. Dataset. 777:777. https://doi.org/10.6084/m9.figshare.880756.v5
Nishiwaki-Matsushima R, Ohta T, Nishiwaki S, Suganuma M, Kohyama K, Ishikawa T, Carmichael WW, Fujiki H (1992) Liver tumor promotion by the cyanobacterial cyclic peptide toxin microcystin-LR. J Cancer Res Clin Oncol 118:420–424. https://doi.org/10.1007/BF01629424
Ohta T, Sueoka E, Iida N, Komori A, Suganuma M, Nishiwaki R, Tatematsu M, Kim S-J, Carmichael WW, Fujiki H (1994) Nodularin, a potent inhibitor of protein phosphatases 1 and 2A, is a new environmental carcinogen in male F344 rat liver. Cancer Res 54:6402–6406
Runnegar M, Berndt N, Kong SM, Lee EY, Zhang L (1995) In vivo and in vitro binding of microcystin to protein phosphatases 1 and 2A. Biochem Biophys Res Commun 216:162–169. https://doi.org/10.1006/bbrc.1995.2605
Sedan D, Laguens M, Copparoni G, Aranda JO, Giannuzzi L, Marra CA, Andrinolo D (2015) Hepatic and intestine alterations in mice after prolonged exposure to low oral doses of microcystin-LR. Toxicon 104:26–33. https://doi.org/10.1016/j.toxicon.2015.07.011
Sekijima M, Tsutsumi T, Yoshida T, Harada T, Tashiro F, Chen G, Yu S-Z, Ueno Y (1999) Enhancement of glutathione S-transferase placental-form positive liver cell foci development by microcystin-LR in aflatoxin B1-initiated rats. Carcinogenesis 20:161–165. https://doi.org/10.1093/carcin/20.1.161
Sumner LW, Amberg A, Barrett D, Beale MH, Beger R, Daykin CA, Fan TW-M, Fiehn O, Goodacre R, Griffin JL, Hankemeier T, Hardy N, Harnly J, Higashi R, Kopka J, Lane AN, Lindon JC, Marriott P, Nicholls AW, Thaden JJ, Viant MR (2007) Proposed minimum reporting standards for chemical analysis. Metabolomics 3:211–221. https://doi.org/10.1007/s11306-007-0082-2
Ueno Y, Nagata S, Tsutsumi T, Hasegawa A, Watanabe MF, Park H-D, Chen G-C, Chen G, Yu S-Z (1996) Detection of microcystins, a blue-green algal hepatotoxin, in drinking water sampled in Haimen and Fusui, endemic areas of primary liver cancer in China, by highly sensitive immunoassay. Carcinogenesis 17:1317–1321
Ueno Y, Makita Y, Nagata S, Tsutsumi T, Yoshida F, Tamura S-I, Sekijima M, Tashiro F, Harada T, Yoshida T (1999) No chronic oral toxicity of a low dose of microcystin-LR, a cyanobacterial hepatotoxin, in female BALB/c mice. Environ Toxicol 14:45–55. https://doi.org/10.1002/(SICI)1522-7278(199902)14:1%3c45:AID-TOX8%3e3.0.CO;2-T
van Apeldoorn ME, van Egmond HP, Speijers GJA, Bakker GJI (2007) Toxins of cyanobacteria. Mol Nutr Food Res 51:7–60. https://doi.org/10.1002/mnfr.200600185
Worley B, Powers R (2013) Multivariate analysis in metabolomics. Curr. Metabolomics 1:92–107. https://doi.org/10.2174/2213235X11301010092
Worley B, Powers R (2016) PCA as a practical indicator of OPLS-DA model reliability. Curr Metabolomics 4:97–103. https://doi.org/10.2174/2213235X04666160613122429
Zegura B (2016) An overview of the mechanisms of microcystin-LR genotoxicity and potential carcinogenicity. Mini-Rev Med Chem 16:1042–1062. https://doi.org/10.2174/1389557516666160308141549
Zhang Z, Zhang X-X, Wu B, Yin J, Yu Y, Yang L (2016) Comprehensive insights into microcystin-LR effects on hepatic lipid metabolism using cross-omics technologies. J Hazard Mater 315:126–134. https://doi.org/10.1016/j.jhazmat.2016.05.011
Zhao Y, Xie P, Fan H (2012) Genomic profiling of microRNAs and proteomics reveals an early molecular alteration associated with tumorigenesis induced by MC-LR in mice. Environ Sci Technol 46:34–41. https://doi.org/10.1021/es201514h
Zhao S, Li G, Chen J (2015) A proteomic analysis of prenatal transfer of microcystin-LR induced neurotoxicity in rat offspring. J Proteomics 114:197–213. https://doi.org/10.1016/j.jprot.2014.11.015
Zhao S, Xie P, Chen J, Liu L, Fan H (2016) A proteomic study on liver impairment in rat pups induced by maternal microcystin-LR exposure. Environ Pollut 212:197–207. https://doi.org/10.1016/j.envpol.2015.12.055
Zhou M, Tu W, Xu J (2015) Mechanisms of microcystin-LR-induced cytoskeletal disruption in animal cells. Toxicon 101:92–100. https://doi.org/10.1016/j.toxicon.2015.05.005
Acknowledgements
The authors would like to thank the Centre for Public Health at Queen’s University Belfast (UK) for the blood biochemistry analysis using the RX Daytona + clinical chemistry analyzer (Randox Laboratories, UK). We would also like to thank the Veterinary Sciences Division, Agri-Food and Biosciences Institute, Belfast, Northern Ireland for undertaking the animal experiments.
Funding
This work was supported by the Science Foundation Ireland (SFI)-DEL Investigators Programme Partnership (Project 14/IA/2646) and the European Union Horizon 2020 Research and Innovation Programme under Grant Agreement No. 692195 (‘MultiCoop’).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Research Involving Animals: Ethical Approval
The animal experiment was performed under license (PPL 2755—Chemical contaminants in food producing animals) issued by the Department of Health, Social Services and Public Safety in Northern Ireland (NI) in accordance with the Animal (Scientific Procedures) Act (1986), and under the approval of the Agri-Food and Biosciences Institute (AFBI) Animal Welfare and Ethical Review Body (AWERB) and associated EU Directive 2010/63/EU for animal experiments. The study was carried out on behalf of Queen’s University Belfast by the Veterinary Sciences Division, AFBI, Belfast, Northern Ireland.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Welten, R.D., Meneely, J.P., Chevallier, O.P. et al. Oral Microcystin-LR Does Not Cause Hepatotoxicity in Pigs: Is the Risk of Microcystin-LR Overestimated?. Expo Health 12, 775–792 (2020). https://doi.org/10.1007/s12403-019-00336-6
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12403-019-00336-6