Abstract
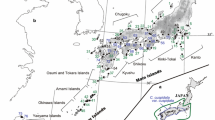
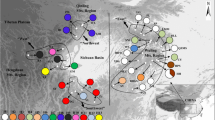
Of the nearly 40 endemics present on Ulleung Island, Campanula takesimana Nakai represents an anagenetically derived lineage of a continental progenitor, C. punctata Lam. Other than its low morphological divergence, little is known about the genetic diversity and population structure of the species pair or the geographical origin of C. takesimana. We sampled a total of 240 accessions in 22 populations, including one Dokdo Island population, of the two species, sequenced four noncoding chloroplast regions (rps16-trnK, trnQ-rps16, psbD-trnT, and psbM-trnD; 4482 bp), and assessed the genetic consequences of anagenetic speciation. Based on chloroplast DNA, we found substantially lower genetic diversity statistics for C. takesimana compared to its progenitor, significant population genetic structuring in insular derivative species, and significant molecular divergence between C. punctata and C. takesimana. Mutually exclusive haplotypes were found in the two species, and the haplotype network suggested that Ulleung Island haplotypes were derived from the Dokdo Island haplotype, which was originally derived from a C. punctata population in Bonghwa-gun, Gyeongsangbuk-do Province. This study pinpoints a very narrow geographical source area and suggests the potentially important role of Dokdo Island as an initial stepping stone for Ulleung Island endemics.




Similar content being viewed by others
References
Birky CW (1995) Uniparental inheritance of mitochondrial and chloroplast genes: mechanisms and evolution. Proc Natl Acad Sci USA 92:11331–11338
Clement M, Posada D, Crandall KA (2000) TCS a computer program to estimate gene genealogies. Mol Ecol 9:1657–1659
Dupanloup I, Schneider S, Excoffier L (2002) A simulated annealing approach to define the genetic structure of populations. Mol Ecol 11:2571–2581
Emerson BC, Patiño J (2018) Anagenesis, cladogenesis, and speciation on islands. Trends Ecol Evol 33:488–491
Excoffier L, Lischer HEL (2010) Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Mol Ecol Resour 10:564–567
Fu YX (1997) Statistical tests of neutrality of mutations against population growth, hitchhiking and background selection. Genetics 147:915–925
Gil H-Y, Maki M, Pimenova EA, Taran A, Kim S-C (2020) Origin of the critically endangered endemic species Scrophularia takesimensis (Scrophulariaceae) on Ulleung Island, Korea: implications for conservation. J Plant Res. https://doi.org/10.1007/s10265-020-01221-z
Hare MP (2001) Prospects for nuclear gene phylogeography. Trends Ecol Evol 16:700–706
Harpending HC (1994) Signature of ancient population growth in a low-resolution mitochondrial DNA mismatch distribution. Hum Biol 66:591–600
Hart MW, Sunday J (2007) Things fall apart: biological species form unconnected parsimony networks. Biol Lett 3:509–512
Havill N, Campbell CS, Vining T, LePage B, Bayer R, Donoghue M (2008) Phylogeny and biogeography of Tsuga (Pinaceae) inferred from nuclear ribosomal ITS and chloroplast DNA sequence data. Syst Bot 33:478–489
Holman G, Del Tredici P, Havill N, Lee NS, Cronn R, Cushman K, Mathews S, Raubeson L, Campbell CS (2017) A new species and introgression in eastern Asian Hemlocks (Pinaceae: Tsuga). Syst Bot 42:733–746
Hyun JO, Kwon SK (2006) Flora of Dokdo, report on the detailed survey of Dokdo ecosystem. Ministry of Environment, Seoul, pp 35–44
Kim YK (1985) Petrology of ulreung volcanic island, Korea—part 1. Geology. J Jpn Assoc Mineral Petrol Econ Geol 80:128–135
Kim JW (1988) The phytosociology of forest vegetation on Ulreung-do, Korea. Phytocoenologia 16:128–135
Kim K-A, Yoo K-O (2012) Phylogenetic relationships of Korean Campanulaceae based on chloroplast DNA sequences. Korean J Pl Taxon 42:282–293
Kim HJ, Shin BC, Jeung MY (2009) Research trend of Dokdo island plants-at focus of research data from 1947 to 2009. J Korean Inst Tradit Landsc Archit 27:61–78
Kim K-A, Yoo K-O, Cheon K-S (2020) The complete chloroplast genome sequence of Campanula zangezura (Campanulaceae). Mitochondrial DNA B Resour 5:480–481
Lee W, Pak J-H (2010) Intraspecific sequence variation of trnL/F intergenic region (cpDNA) in Sedum takesimense Nakai (Crassulaceae) and aspects of geographic distribution. Korean J Pl Taxon 40:157–162
Lee S, Lee J, Kim S (1997) A new species of Adenophora (Campanulaceae) from Korea. J Plant Res 110:77–80
Lee W, Yang JY, Jung K-S, Pak J-H, Maki M, Kim S-C (2017) Chloroplast DNA assessment of anagenetic speciation in Rubus takesimensis (Rosaceae) on Ulleung Island, Korea. J Plant Biol 60:163–174
Librado P, Rozas J (2009) DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25:1451–1452
Maddison WP, Maddison DR (2005) MacClade version 4.08 Analysis of phylogeny and character evolution. Sinauer Associates, Sunderland
Muller K (2005) SeqState—primer design and sequence statistics for phylogenetic DNA data sets. Appl Bioinform 4:65–69
Oh S-H (2015) Sea, wind, or bird: origin of Fagus multinervis (Fagaceae) inferred from chloroplast DNA sequences. Korean J Pl Taxon 45:213–220
Palumbi SR, Cipriano F, Hare MP (2001) Predicting nuclear gene coalescence from mitochondrial data: the three-times rule. Evolution 55:859–868
Park K-R, Jung H-J (2000) Isozyme and morphological variation in Campanula punctata and C. takesimina (Campanulaceae). Korean J Pl Taxon 30:1–16
Park C-W, Bhandari GS, Won H, Park JH, Park DS (2018) Polyploidy and introgression in invasive giant knotweed (Fallopia sachalinensis) during the colonization of remote volcanic islands. Sci Rep 8:16021
Pfosser MF, Guzy-Wróbelska J, Sun B-Y, Stuessy TF, Sugawara T, Jujii N (2002) The origin of species of Acer (Sapindaceae) endemic to Ullung Island, Korea. Syst Bot 27:351–367
Pfosser MF, Jakubowsky G, Schluter PM, Fer T, Kato H, Stuessy TF, Sun BY (2005) Evolution of Dystaenia takesimana (Apiaceae), endemic to Ulleung Island, Korea. Plant Syst Evol 256:159–170
Rogers AR, Hapending H (1992) Population growth makes waves in the distribution of pairwise genetic differences. Mol Biol Evol 9:552–569
Roquet C, Sanmartín I, Garcia-Jacas N, Sáez L, Susanna A, Wikström N, Aldasoro JJ (2009) Reconstructing the history of Campanulaceae with a Bayesian approach to molecular dating and dispersal-vicariance analyses. Mol Phylogenet Evol 52:575–587
Schneider S, Excoffier L (1999) Estimation of past demographic parameters from the distribution of pairwise differences when the mutation rates vary among sites: application to human mitochondrial DNA. Genetics 152:1079–1089
Seo H-S (2018) Population structure of Phedimus takesimensis (Crassulaceae) on Ulleung Island and molecular comparison with P. kamtschaticus, a continental progenitor. MS Thesis, Sungkyunkwan University, Korea
Shaw J, Lickey EB, Schilling EE, Small RL (2007) Comparison of whole chloroplast genome sequences to choose noncoding regions for phylogenetic studies in angiosperms: The tortoise and the hare III. Am J Bot 94:275–288
Shetler SG, Morin N (1986) Seed morphology in North American Campanulaceae. Ann Missouri Bot Gard 73:653–688
Simmon MP, Ochoterena H (2000) Gaps as characters in sequence-based phylogenetic analyses. Syst Biol 49:369–381
Slatkin M, Hudson RR (1991) Pairwise comparisons of mitochondrial DNA sequences in stable and exponentially growing populations. Genetics 129:555–562
Sohn YK, Park KH (1994) Geology and evolution of Tok Island, Korea. J Geol Soc Korea 3:242–261
Stevens PF (2001 onward) Angiosperm Phylogeny Website. Version 14, July 2017. https://www.mobot.org/MOBOT/research/APweb/
Stuessy TF, Ono M (1998) Evolution and speciation of island plants. Cambridge University Press, New York
Stuessy TF, Jakubowsky G, Gómez RS, Pfosser M, Schlüter PM, Fer T, Sun B-Y, Kato H (2006) Anagenetic evolution in island plants. J Biogeogr 33:1259–1265
Stuessy TF, Takayama K, López-Sepúlveda P, Crawford DJ (2014) Interpretation of patterns of genetic variation in endemic plant species of oceanic islands. Bot J Linn Soc 174:276–288
Sun B-Y, Stuessy TF (1998) Preliminary observations on the evolution of endemic angiosperms of Ullung Island, Korea. In: Stuessy TF, Ono M (eds) Evolution and speciation of island plants. Cambridge University Press, New York, pp 181–202
Sun B-Y, Sul MR, Im JA, Kim CH, Kim TJ (2002) Evolution of endemic vascular plants of Ulleungdo and Dokdo in Korea-floristic and cytotaxonomic characteristics of vascular flora of Dokdo. Korean J Pl Taxon 32:143–158
Sun B-Y, Shin H, Hyun J-O, Kim Y-D, Oh S-H (2014) Vascular plants of Dokdo and Ulleungdo Islands in Korea. The National Institute of Biological Resources, GeoBook Publishing Co, Korea
Tackenberg O, Poschlod P, Bonn S (2003) Assessment of wind dispersal potential in plant species. Ecol Monogr 73:191–205
Tajima F (1989) Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 123:585–595
Takayama K, Sun B-Y, Stuessy TF (2012) Genetic consequences of anagenetic speciation in Acer okamotoanum (Sapindaceae) on Ullung Island, Korea. Ann Bot 109:321–330
Takayama K, Sun B-Y, Stuessy TF (2013) Anagenetic speciation in Ullung Island, Korea: genetic diversity and structure in the island endemic species, Acer takesimense (Sapindaceae). J Plant Res 126:323–333
Takayama K, López-Sepúlveda P, Greimler J, Crawford DJ, Peñailillo P, Baeza M, Ruiz E, Kohl G, Tremetsberger K, Gatica A, Letelier L, Novoa P, Novak J, Stuessy TF (2015a) Genetic consequences of cladogenetic vs anagenetic speciation in endemic plants on oceanic islands. AoB Plants 7:plv102. https://doi.org/10.1093/aobpla/pvl102
Takayama K, López-Sepúlveda P, Greimler J, Crawford DJ, Peñailillo P, Baeza M, Ruiz E, Kohl G, Tremetsberger K, Gatica A, Letelier L, Novoa P, Novak J, Stuessy TF (2015b) Relationships and genetic cosequences of contrasting modes of speciation among endemic species of Robinsonia (Asteraceae, Senecioneae) of the Juan Fernandez Archipelago, Chile, based on AFLPs and SSRs. New Phytol 205:415–428
Trusenkova OO, Stanichnyi SV, Ratner YB (2007) Major variability modes and wind patterns over the Sea of Japan and adjacent areas. Izv Atmos Ocean Phys 43:688–703
Weiss H, Sun B-Y, Stuessy TF, Kim C-H, Kato H, Wakabayashi M (2002) Karyology of plant species endemic to Ulleung Island (Korea) and selected relatives in peninsular Korea and Japan. Bot J Linn Soc 138:93–105
Yang J, Pak J-H, Maki M, Kim S-C (2019) Multiple origins and the population genetic structure of Rubus takesimensis (Rosaceae) on Ulleng Island: implications for the genetic consequences of anagenetic speciation. PLoS ONE 14:e0222707
Zink RM, Barrowclough GF (2008) Mitochondrial DNA under siege in avian phylogeography. Mol Ecol 17:2107–2121
Acknowledgements
We thank two anonymous reviewers for their helpful comments and suggestions on an earlier version of the manuscript. This research was supported by the National Research Foundation of Korea (NRF-2016R1D1A2B03934596 and NRF-2019R1A2C2009841), to SCK.
Author information
Authors and Affiliations
Contributions
WYC, SHK, JYY, and WL collected samples, and WYC and SHK generated and analyzed the data. JHP and SCK conceived the ideas and provided the overall direction for this project. The initial draft of this manuscript was written by WYC and subsequently edited by SCK. All authors read and approved the final manuscript.
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Cheong, W.Y., Kim, SH., Yang, J. et al. Insights from Chloroplast DNA into the Progenitor-Derivative Relationship Between Campanula punctata and C. takesimana (Campanulaceae) in Korea. J. Plant Biol. 63, 431–444 (2020). https://doi.org/10.1007/s12374-020-09281-3
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12374-020-09281-3