Abstract
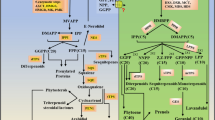
Rice, as a major crop in the world, produces huge agronomic biomass residues besides food, which consist of cellulose, hemicelluloses and lignin. Many master regulators of secondary wall synthesis were identified in the model plant Arabidopsis. In this study, we investigated the function of a NAC (NAM, ATAF, and CUC2) transcription factor related to secondary cell wall biosynthesis, which is highly expressed in rice sclerenchyma tissue and is named OsSWN1. Our results showed that engineering of OsSWN1 could exhibit multiple features regulated to agronomic traits and bioenergy research. Over-expression of OsSWN1 caused an erect-leaf and enclosed-flower phenotype. Secondary cell wall-related genes were actively expressed in transgenic plants with obvious ectopic lignin deposition in the leaf collar, while increased lignin content and decreased the sugar yield correspondingly. In addition, down-regulation of OsSWN1 expression levels decreased lignin content and increased the sugar yield in transgenic plants. Bioinformatics analysis revealed that OsSWN1-like genes are highly conserved in switchgrass and sorghum, suggesting a possibility of manipulating the expression level of the OsSWN1 orthologs in the bioenergy crops for biofuel production.
Similar content being viewed by others
References
Chen F, Dixon RA (2007) Lignin modification improves fermentable sugar yields for biofuel production. Nat Biotechnol 25:759–761
Chen S, Songkumarn P, Liu J, Wang GL (2009) A versatile zero background T-vector system for gene cloning and functional genomics. Plant Physiol 150:1111–1121
Jiao Y, Tausta SL, Gandotra N, Sun N, Liu T, Clay NK, Ceserani T, Chen M, Ma L, Holford M, Zhang H, Zhao H, Deng XW, Nelson T (2009) A transcriptome atlas of rice cell types uncovers cellular, functional and developmental hierarchies. Nat Genet 41:258–263
Kim S, and Dale, B. E. (2003) Blobal potential bioethanol production from wasted crops and crop residues. Biomass Bioenergy 26:361–375
Ko JH, Kim WC, Han KH (2009) Ectopic expression of MYB46 identifies transcriptional regulatory genes involved in secondary wall biosynthesis in Arabidopsis. Plant J 60:649–665
Kosugi S, Suzuka I, Ohashi Y, Murakami T, Arai Y (1991) Upstream sequences of rice proliferating cell nuclear antigen (PCNA) gene mediate expression of PCNA-GUS chimeric gene in meristems of transgenic tobacco plants. Nucleic Acids Res 19:1571–1576
Li Y, Qian Q, Zhou Y, Yan M, Suna L, Zhang M, Fu Z, Wang Y, Han B, Pang X, Chen M, Li J (2003) BRITTLE CULM1, which encodes a COBRA-like protein, affects the mechanical properties of rice plants. Plant Cell 15:2020–2031
McCarthy RL, Zhong R, Ye ZH (2009) MYB83 is a direct target of SND1 and acts redundantly with MYB46 in the regulation of secondary cell wall biosynthesis in Arabidopsis. Plant Cell Physiol 50:1950–1964
Miki D, Itoh R, Shimamoto K (2005) RNA silencing of single and multiple members in a gene family of rice. Plant Physiol 138:1903–1913
Mitsuda N, Iwase A, Yamamoto H, Yoshida M, Seki M, Shinozaki K, Ohme-Takagi M (2007) NAC transcription factors, NST1 and NST3, are key regulators of the formation of secondary walls in woody tissues of Arabidopsis. Plant Cell 19:270–280
Mitsuda N, Seki M, Shinozaki K, Ohme-Takagi M (2005) The NAC transcription factors NST1 and NST2 of Arabidopsis regulate secondary wall thickenings and are required for anther dehiscence. Plant Cell 17:2993–3006
Morinaka Y, Sakamoto T, Inukai Y, Agetsuma M, Kitano H, Ashikari M, Matsuoka M (2006) Morphological alteration caused by brassinosteroid insensitivity increases the biomass and grain production of rice. Plant Physiol 141:924–931
Nuruzzaman M, Manimekalai R, Sharoni AM, Satoh K, Kondoh H, Ooka H, Kikuchi S (2010) Genome-wide analysis of NAC transcription factor family in rice. Gene 465:30–44
Qu S, Liu G, Zhou B, Bellizzi M, Zeng L, Dai L, Han B, Wang GL (2006) The broad-spectrum blast resistance gene Pi9 encodes a nucleotide-binding site-leucine-rich repeat protein and is a member of a multigene family in rice. Genetics 172:1901–1914
Sakamoto T, Morinaka Y, Ohnishi T, Sunohara H, Fujioka S, Ueguchi-Tanaka M, Mizutani M, Sakata K, Takatsuto S, Yoshida S, Tanaka H, Kitano H, Matsuoka M (2006) Erect leaves caused by brassinosteroid deficiency increase biomass production and grain yield in rice. Nat Biotechnol 24:105–109
Sampedro J, Cosgrove DJ (2005) The expansin superfamily. Genome Biol 6:242
Saxena IM, Brown RM, Jr. (2005) Cellulose biosynthesis: current views and evolving concepts. Ann Bot 96:9–21
Shen H, Yin Y, Chen, F. X, Y., Dixon RA (2009) A bioinformatic analysis of NAC genes for plant cell wall development in relation to lignocellulosic bioenergy production. Bioenergy Res:217–232
Somerville C (2006) Cellulose synthesis in higher plants. Annu Rev Cell Dev Biol 22:53–78
Somerville C (2007) Biofuels. Curr Biol 17:R115–119
Somerville C, Bauer S, Brininstool G, Facette M, Hamann T, Milne J, Osborne E, Paredez A, Persson S, Raab T, Vorwerk S, Youngs H (2004) Toward a systems approach to understanding plant cell walls. Science 306:2206–2211
Tanaka A, Nakagawa H, Tomita C, Shimatani Z, Ohtake M, Nomura T, Jiang CJ, Dubouzet JG, Kikuchi S, Sekimoto H, Yokota T, Asami T, Kamakura T, Mori M (2009) BRASSINOSTEROID UPREGULATED1, encoding a helix-loop-helix protein, is a novel gene involved in brassinosteroid signaling and controls bending of the lamina joint in rice. Plant Physiol 151:669–680
Tanaka K, Murata K, Yamazaki M, Onosato K, Miyao A, Hirochika H (2003) Three distinct rice cellulose synthase catalytic subunit genes required for cellulose synthesis in the secondary wall. Plant Physiol 133:73–83
Toki S, Hara N, Ono K, Onodera H, Tagiri A, Oka S, Tanaka H (2006) Early infection of scutellum tissue with Agrobacterium allows high-speed transformation of rice. Plant J 47:969–976
Turner SR, Taylor N, Jones L (2001) Mutations of the secondary cell wall. Plant Mol Biol 47:209–219
Wang L, Guo K, Li Y, Tu Y, Hu H, Wang B, Cui X, Peng L (2010) Expression profiling and integrative analysis of the CESA/CSL superfamily in rice. BMC Plant Biol 10:282
Yamamuro C, Ihara Y, Wu X, Noguchi T, Fujioka S, Takatsuto S, Ashikari M, Kitano H, Matsuoka M (2000) Loss of function of a rice brassinosteroid insensitive 1 homolog prevents internode elongation and bending of the lamina joint. Plant Cell 12:1591–1606
Yan C, Yan S, Zeng X, Zhang Z, Gu M (2007) Fine mapping and isolation of Bc7(t), allelic to OsCesA4. J Genet Genomics 34:1019–1027
Yokoyama R, Rose JK, Nishitani K (2004) A surprising diversity and abundance of xyloglucan endotransglucosylase/hydrolases in rice. Classification and expression analysis. Plant Physiol 134:1088–1099
Yoshida K, Sakamoto S, Kawai T, Kobayashi Y, Sato K, Ichinose Y, Yaoi K, Akiyoshi-Endo M, Sato H, Takamizo T, Ohme-Takagi M, Mitsuda N (2013) Engineering the Oryza sativa cell wall with rice NAC transcription factors regulating secondary wall formation. Front Plant Sci 4:383–389
Zhang K, Qian Q, Huang Z, Wang Y, Li M, Hong L, Zeng D, Gu M, Chu C, Cheng Z (2006) GOLD HULL AND INTERNODE2 encodes a primarily multifunctional cinnamyl-alcohol dehydrogenase in rice. Plant Physiol 140:972–983
Zhao Q, Gallego-Giraldo L, Wang H, Zeng Y, Ding SY, Chen F, Dixon RA An NAC transcription factor orchestrates multiple features of cell wall development in Medicago truncatula. Plant J 63:100–114
Zhong R, Demura T, Ye ZH (2006) SND1, a NAC domain transcription factor, is a key regulator of secondary wall synthesis in fibers of Arabidopsis. Plant Cell 18:3158–3170
Zhong R, Lee C, McCarthy RL, Reeves CK, Jones EG, Ye ZH (2011a) Transcriptional activation of secondary wall biosynthesis by rice and maize NAC and MYB transcription factors. Plant Cell Physiol 52:1856–1871
Zhong R, Lee C, Zhou J, McCarthy RL, Ye ZH (2008) A battery of transcription factors involved in the regulation of secondary cell wall biosynthesis in Arabidopsis. Plant Cell 20:2763–2782
Zhong R, McCarthy RL, Lee C, Ye ZH (2011b) Dissection of the transcriptional program regulating secondary wall biosynthesis during wood formation in poplar. Plant Physiol 157:1452–1468
Zhong R, Richardson EA, Ye ZH (2007a) The MYB46 transcription factor is a direct target of SND1 and regulates secondary wall biosynthesis in Arabidopsis. Plant Cell 19:2776–2792
Zhong R, Richardson EA, Ye ZH (2007b) Two NAC domain transcription factors, SND1 and NST1, function redundantly in regulation of secondary wall synthesis in fibers of Arabidopsis. Planta 225:1603–1611
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Rights and permissions
About this article
Cite this article
Chai, M., Bellizzi, M., Wan, C. et al. The NAC transcription factor OsSWN1 regulates secondary cell wall development in Oryza sativa . J. Plant Biol. 58, 44–51 (2015). https://doi.org/10.1007/s12374-014-0400-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12374-014-0400-y