Abstract
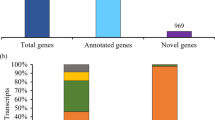
Oryza longistaminata, a perennial wild rice species with an AA genome, is characterized by the presence of rhizomatous stems. The rhizomatous trait in rice was previously shown to be quantitatively controlled by many genes, but the molecular mechanism related to rhizome development is still unknown. In the present study, expressed sequence tags (ESTs) generated from rhizome tips of O. longistaminata were collected and analyzed. A total of 10,283 complimentary deoxyribunucleic acid clones were randomly sequenced, which generated 10,136 raw sequences, and finally, 4,419 unisequences with diverse functional categories were generated. These unisequences were mapped onto the Oryza sativa genome, which revealed that 4,285 (96.97%) and 4,151 (93.94%) of the unisequences were alignable to the japonica and indica genomic sequences, respectively, with >80% sequence identity. Additionally, 41 unisequences showed four typical types of alternative splicing patterns. More than 600 simple sequence repeats were identified in these unisequences. A subset of unisequences were physically colocalized onto rhizome-related quantitative trait locus intervals in rice and sorghum, and one gene, OLRR1, was further confirmed to be enriched in the rhizome tip and young leaf by real-time polymerase chain reaction and in situ hybridization. Unisequences reported in this study provide valuable data for molecular dissection of the rhizomatous growth habit in O. longistaminata.




Similar content being viewed by others
References
Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25:3389–3402
Black DL (2003) Mechanisms of alternative pre-messenger RNA splicing. Annu Rev Biochem 72:291–336
Blencowe BJ (2006) Alternative splicing: new insights from global analyses. Cell 126:37–47
Cho SK, Ok SH, Jeung JU, Shim KS, Jung KW, You MK, Kang KH, Chung YS, Choi HC, Moon HP, Shin JS (2004) Comparative analysis of 5,211 leaf ESTs of wild rice (Oryza minuta). Plant Cell Rep 22:839–847
Dello Ioio R, Nakamura K, Moubayidin L, Perilli S, Taniguchi M, Morita MT, Aoyama T, Costantino P, Sabatini S (2008) A genetic framework for the control of cell division and differentiation in the root meristem. Science 322:1380–1384
Fahlgren N, Montgomery TA, Howell MD, Allen E, Dvorak SK, Alexander AL, Carrington JC (2006) Regulation of AUXIN RESPONSE FACTOR3 by TAS3 ta-siRNA affects developmental timing and patterning in Arabidopsis. Curr Biol 16:939–944
Ghesquiere A (1985) Evolution of Oryza longistaminata. In: Stephen JB (ed) Rice genetics I. International Rice Research Institute (IRRI), Philippines, pp 15–27
Ghesquiere A (1991) Re-examination of genetic control of the reproductive barrier between Oryza longistaminata and O. sativa and relationship with rhizome expression. In: Khush GS (ed) Rice genetics II. International Rice Research Institute (IRRI), Philippines, pp 729–730
Ghesquiere A, Causse M (1992) Linkage study between molecular markers and genes controlling the reproductive barrier in interspecific backcross between O. sativa and O. longistaminata. RGN 9:28–31
Gomez LD, Gilday A, Feil R, Lunn JE, Graham IA (2010) AtTPS1-mediated trehalose 6-phosphate synthesis is essential for embryogenic and vegetative growth and responsiveness to ABA in germinating seeds and stomatal guard cells. Plant J 64:1–13
Graveley BR (2001) Alternative splicing: increasing diversity in the proteomic world. Trends Genet 17:100–107
Hu FY, Tao DY, Sacks E, Fu BY, Xu P, Li J, Yang Y, McNally K, Khush GS, Paterson AH, Li ZK (2003) Convergent evolution of perenniality in rice and sorghum. Proc Natl Acad Sci USA 100:4050–4054
Hu F, Wang D, Zhao X, Zhang T, Sun H, Zhu L, Zhang F, Li L, Li Q, Tao D, Fu B, Li Z (2011) Identification of rhizome-specific genes by genome-wide differential expression Analysis in Oryza longistaminata. BMC Plant Biol 11:18
International Rice Genome Sequencing Project (2005) The map-based sequence of the rice genome. Nature 436:793–800
Jackson D (1991) In-situ hybridization in plants. In: Bowles DJ, Gurr SJ, McPherson M (eds) Molecular plant pathology: a practical approach. Oxford Univ, Press, Oxford, UK, pp 163–174
Jang CS, Kamps TL, Skinner DN, Schulze SR, Vencill WK, Paterson AH (2006) Functional classification, genomic organization, putatively cis-acting regulatory elements, and relationship to quantitative trait loci, of sorghum genes with rhizome-enriched expression. Plant Physiol 142:1148–1159
Jang CS, Kamps TL, Tang H, Bowers JE, Lemke C, Paterson AH (2009) Evolutionary fate of rhizome-specific genes in a non-rhizomatous sorghum genotype. Heredity 102:266–273
Kent WJ (2002) BLAT—the BLAST-like alignment tool. Genome Res 12:656–664
Lu TT, Yu S, Fan D, Mu J, Shangguan Y, Wang Z, Minobe Y, Lin Z, Han B (2008) Collection and comparative analysis of 1888 full-length cDNAs from wild rice Oryza rufipogon Griff. W1943. DNA Res 15:285–295
Lu T, Lu G, Fan D, Zhu C, Li W, Zhao Q, Feng Q, Zhao Y, Guo Y, Huang X, Han B (2010) Function annotation of the rice transcriptome at single-nucleotide resolution by RNA-seq. Genome Res 20:1238–1249
Maekawa M, Inukai T, Rikiishi K, Matsuura T, Govidaraj KG (1998) Inheritance of the rhizomatous traits in hybrid of Oryza longistaminata Chev. et Roehr. and O. sativa L. SABRAO. J Breeding Genet 30:69–72
Nakagahra M, Okuno K, Vaughan D (1997) Rice genetic resources history, conservation, investigative characterization and use in Japan. Plant Mol Biol 35:69–77
Nicot N, Chiquet V, Gandon B, Amilhat L, Legeai F, Leroy P, Bernard M, Sourdille P (2004) Study of simple sequence repeat (SSR) markers from wheat expressed sequence tags (ESTs). Theor Appl Genet 109:800–805
Park MY, Chung MS, Koh HS, Lee DJ, Ahn SJ, Kim CS (2009) Isolation and functional characterization of the Arabidopsis salt-tolerance 32 (AtSAT32) gene associated with salt tolerance and ABA signaling. Physiol Plant 135:426–435
Paterson AH, Schertz KF, Lin YR, Liu SC, Chang YL (1995) The weediness of wild plants: molecular analysis of genes influencing dispersal and persistence of johnsongrass, Sorghum halepense (L.) Pers. Proc Natl Acad Sci U S A 92:6127–6131
Paterson AH, Bowers JE, Bruggmann R, Dubchak I, Grimwood J, Gundlach H, Haberer G, Hellsten U, Mitros T, Poliakov A, Schmutz J, Spannagl M, Tang H, Wang X, Wicker T, Bharti AK, Chapman J, Feltus FA, Gowik U, Grigoriev IV, Lyons E, Maher CA, Martis M, Narechania A, Otillar RP, Penning BW, Salamov AA, Wang Y, Zhang L, Carpita NC, Freeling M, Gingle AR, Hash CT, Keller B, Klein P, Kresovich S, McCann MC, Ming R, Peterson DG, Mehboob ur R, Ware D, Westhoff P, Mayer KFX, Messing J, Rokhsar DS (2009) The Sorghum bicolor genome and the diversification of grasses. Nature 457:551–556
Reddy AS (2007) Alternative splicing of pre-messenger RNAs in plants in the genomic era. Annu Rev Plant Biol 58:267–294
Sacks EJ, Roxas JP, Sta CM (2003) Developing perennial upland rice II: filed performance of S1 families from an intermated Oryza sativa/O. longistaminata population. Crop Sci 43:129–134
Song WY, Wang GL, Chen LL, Kim HS, Pi LY, Holsten T, Gardner J, Wang B, Zhai WX, Zhu LH, Fauquet C, Ronald P (1995) A receptor kinase-like protein encoded by the rice disease resistance gene, Xa21. Science 270:1804–1806
Tanksley SD, McCouch SR (1997) Seed banks and molecular maps: unlocking genetic potential from the wild. Science 277:1063–1066
Thiel T, Michalek W, Varshney RK, Graner A (2003) Exploiting EST databases for the development and characterization of gene-derived SSR-markers in barley (Hordeum vulgare L.). Theor Appl Genet 106:411–422
Vaughan DA (1994) Wild relatives of rice: genetic resources handbook. International Rice Research Institute (IRRI), Philippines, pp 46–47
Wang K, Peng H, Lin E, Jin Q, Hua X, Yao S, Bian H, Han N, Pan J, Wang J, Deng M, Zhu M (2009a) Identification of genes related to the development of bamboo rhizome bud. J Exp Bot 61:551–561
Wang Z, Gerstein M, Snyder M (2009b) RNA-Seq: a revolutionary tool for transcriptomics. Nat Rev Genet 10:57–63
Yamamoto K, Sasaki T (1997) Large-scale EST sequencing in rice. Plant Mol Biol 35:135–144
Yang H, Hu L, Hurek T, Reinhold-Hurek B (2010) Global characterization of the root transcriptome of a wild species of rice, Oryza longistaminata, by deep sequencing. BMC Genomics 11:705
Yu J, Hu S, Wang J, Wong GK, Li S, Liu B, Deng Y, Dai L, Zhou Y, Zhang X, Cao M, Liu J, Sun J, Tang J, Chen Y, Huang X, Lin W, Ye C, Tong W, Cong L, Geng J, Han Y, Li L, Li W, Hu G, Li J, Liu Z, Qi Q, Li T, Wang X, Lu H, Wu T, Zhu M, Ni P, Han H, Dong W, Ren X, Feng X, Cui P, Li X, Wang H, Xu X, Zhai W, Xu Z, Zhang J, He S, Xu J, Zhang K, Zheng X, Dong J, Zeng W, Tao L, Ye J, Tan J, Chen X, He J, Liu D, Tian W, Tian C, Xia H, Bao Q, Li G, Gao H, Cao T, Zhao W, Li P, Chen W, Zhang Y, Hu J, Liu S, Yang J, Zhang G, Xiong Y, Li Z, Mao L, Zhou C, Zhu Z, Chen R, Hao B, Zheng W, Chen S, Guo W, Tao M, Zhu L, Yuan L, Yang H (2002) A draft sequence of the rice genome (Oryza sativa L. ssp. indica). Science 296:79–92
Zeng J, Wang Q, Lin J, Deng K, Zhao X, Tang D, Liu X (2010) Arabidopsis cryptochrome-1 restrains lateral roots growth by inhibiting auxin transport. J Plant Physiol 167:670–673
Zhang J, Feng Q, Jin C, Qiu D, Zhang L, Xie K, Yuan D, Han B, Zhang Q, Wang S (2005) Features of the expressed sequences revealed by a large-scale analysis of ESTs from a normalized cDNA library of the elite indica rice cultivar Minghui 63. Plant J 42:772–780
Zhang G, Guo G, Hu X, Zhang Y, Li Q, Li R, Zhuang R, Lu Z, He Z, Fang X, Chen L, Tian W, Tao Y, Kristiansen K, Zhang X, Li S, Yang H, Wang J (2010) Deep RNA sequencing at single base-pair resolution reveals high complexity of the rice transcriptome. Genome Res 20:646–654
Acknowledgment
This work was supported by the National Natural Science Foundation of China (grant no. U0836605) and the Key Project from MOA (grant no. 2008ZX001-003).
Author information
Authors and Affiliations
Corresponding authors
Electronic supplementary material
Rights and permissions
About this article
Cite this article
Zhang, T., Li, L., Hu, F. et al. Analysis of ESTs from a Normalized cDNA Library of the Rhizome Tip of Oryza longistaminata . J. Plant Biol. 55, 33–42 (2012). https://doi.org/10.1007/s12374-011-9187-2
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12374-011-9187-2