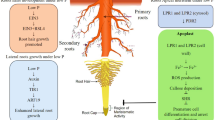
Abstract
The mechanisms of how plants sense nutrient availability in soils and then send the signals through downstream signal cascades are essential processes for plant survival. Although understanding the regulatory mechanisms of macronutrient sensing and signaling are very important, many components still remain unknown. This review describes the recent progress in identifying the transcriptional regulatory components that respond to phosphorus, nitrogen, potassium, and sulfur limitation. Under phosphorus limited conditions, a MYB transcription factor, PHR1, functions as a key transcriptional regulator of the phosphorus deficient responsive genes via binding to the P1BS cis element. Similarly, SLIM1 plays crucial roles in the expression of the sulfur starved responsive genes through binding to the sulfur responsive cis element. The nitrogen deficient responsive MYB transcription factors control nitrogen metabolism. Cytokinins and microRNAs (miRNAs) also mediate macronutrient metabolic processes. Much less is known about the potassium deficient signaling components, as compared to the other nutrient signaling pathways. Multiple recent studies have suggested that phytohormones, such as ethylene, auxin, jasmonic acid, and abscisic acid (ABA), and reactive oxygen species (ROS) play roles as transcriptional regulatory components in potassium deficient signaling.



Similar content being viewed by others
Abbreviations
- bHLH:
-
helix–loop–helix
- miRNA:
-
microRNA
- ROS:
-
reactive oxygen species
References
Amtmann A, Hammond JP, Armengaud P, White PJ (2006) Nutrient sensing and signaling in plants: potassium and phosphorus. Adv Bot Res 43:209–257
Armengaud P, Breitling R, Amtmann A (2004) The potassium-dependent transcriptome of Arabidopsis reveals a prominent role of jasmonic acid in nutrient signaling. Plant Physiol 136:2556–2576
Armengaud P, Sulpice R, Miller AJ, Stitt M, Amtmann A, Gibon Y (2009) Multilevel analysis of primary metabolism provides new insights into the role of potassium nutrition for glycolysis and nitrogen assimilation in Arabidopsis roots. Plant Physiol 150:772–785
Bari R, Datt Pant B, Stitt M, Scheible WR (2006) PHO2, microRNA399, and PHR1 define a phosphate-signaling pathway in plants. Plant Physiol 141:988–999
Baudry A, Heim MA, Dubreucq B, Caboche M, Weisshaar B, Lepiniec L (2004) TT2, TT8, and TTG1 synergistically specify the expression of BANYULS and proanthocyanidin biosynthesis in Arabidopsis thaliana. Plant J 39:366–380
Bi YM, Wang RL, Zhu T, Rothstein SJ (2007) Global transcription profiling reveals differential responses to chronic nitrogen stress and putative nitrogen regulatory components in Arabidopsis. BMC Genomics 8:281
Burleigh SH, Harrison MJ (1997) A novel gene whose expression in Medicago truncatula roots is suppressed in response to colonization by vesicular–arbuscular mycorrhizal (VAM) fungi and to phosphate nutrition. Plant Mol Biol 34:199–208
Calderon-Vazquez C, Ibarra-Laclette E, Caballero-Perez J, Herrera-Estrella L (2008) Transcript profiling of Zea mays roots reveals gene responses to phosphate deficiency at the plant- and species-specific levels. J Exp Bot 59:2479–2497
Castaings L, Camargo A, Pocholle D, Gaudon V, Texier Y, Boutet-Mercey S, Taconnat L, Renou JP, Daniel-Vedele F, Fernandez E, Meyer C, Krapp A (2009) The nodule inception-like protein 7 modulates nitrate sensing and metabolism in Arabidopsis. Plant J 57:426–435
Devaiah BN, Raghothama KG (2007) Transcriptional regulation of Pi starvation responses by WRKY75. Plant Signal Behav 2:424–425
Devaiah BN, Karthikeyan AS, Raghothama KG (2007a) WRKY75 transcription factor is a modulator of phosphate acquisition and root development in Arabidopsis. Plant Physiol 143:1789–1801
Devaiah BN, Nagarajan VK, Raghothama KG (2007b) Phosphate homeostasis and root development in Arabidopsis are synchronized by the zinc finger transcription factor ZAT6. Plant Physiol 145:147–159
Devaiah BN, Madhuvanthi R, Karthikeyan AS, Raghothama KG (2009) Phosphate starvation responses and gibberellic acid biosynthesis are regulated by the MYB62 transcription factor in Arabidopsis. Mol Plant 2:43–58
Falkenberg B, Witt I, Zanor MI, Steinhauser D, Mueller-Roeber B, Hesse H, Hoefgen R (2008) Transcription factors relevant to auxin signaling coordinate broad-spectrum metabolic shifts including sulphur metabolism. J Exp Bot 59:2831–2846
Franco-Zorrilla JM, Martin AC, Solano R, Rubio V, Leyva A, Paz-Ares J (2002) Mutations at CRE1 impair cytokinin-induced repression of phosphate starvation responses in Arabidopsis. Plant J 32:353–360
Franco-Zorrilla JM, Gonzalez E, Bustos R, Linhares F, Leyva A, Paz-Ares J (2004) The transcriptional control of plant responses to phosphate limitation. J Exp Bot 55:285–293
Franco-Zorrilla JM, Martin AC, Leyva A, Paz-Ares J (2005) Interaction between phosphate-starvation, sugar, and cytokinin signaling in Arabidopsis and the roles of cytokinin receptors CRE1/AHK4 and AHK3. Plant Physiol 138:847–857
Gan Y, Filleur S, Rahman A, Gotensparre S, Forde BG (2005) Nutritional regulation of ANR1 and other root-expressed MADS-box genes in Arabidopsis thaliana. Planta 222:730–742
Gifford ML, Dean A, Gutierrez RA, Coruzzi GM, Birnbaum KD (2008) Cell-specific nitrogen responses mediate developmental plasticity. Proc Natl Acad Sci USA 105:803–808
Gutierrez RA, Stokes TL, Thum K, Xu X, Obertello M, Katari MS, Tanurdzic M, Dean A, Nero DC, McClung CR, Coruzzi GM (2008) Systems approach identifies an organic nitrogen-responsive gene network that is regulated by the master clock control gene CCA1. Proc Natl Acad Sci USA 105:4939–4944
Hammond JP, Bennett MJ, Bowen HC, Broadley MR, Eastwood DC, May ST, Rahn C, Swarup R, Woolaway KE, White PJ (2003) Changes in gene expression in Arabidopsis shoots during phosphate starvation and the potential for developing smart plants. Plant Physiol 132:578–596
Hammond JP, Broadley MR, White PJ (2004) Genetic responses to phosphorus deficiency. Ann Bot 94:323–332
Hammond JP, Broadley MR, Craigon DJ, Higgins J, Emmerson ZF, Townsend HJ, White PJ, May ST (2005) Using genomic DNA-based probe-selection to improve the sensitivity of high-density oligonucleotide arrays when applied to heterologous species. Plant Methods 1:10
Hilson P, Allemeersch J, Altmann T, Aubourg S, Avon A, Beynon J, Bhalerao RP, Bitton F, Caboche M, Cannoot B, Chardakov V, Cognet-Holliger C, Colot V, Crowe M, Darimont C, Durinck S, Eickhoff H, de Longevialle AF, Farmer EE, Grant M, Kuiper MT, Lehrach H, Leon C, Leyva A, Lundeberg J, Lurin C, Moreau Y, Nietfeld W, Paz-Ares J, Reymond P, Rouze P, Sandberg G, Segura MD, Serizet C, Tabrett A, Taconnat L, Thareau V, Van Hummelen P, Vercruysse S, Vuylsteke M, Weingartner M, Weisbeek PJ, Wirta V, Wittink FR, Zabeau M, Small I (2004) Versatile gene-specific sequence tags for Arabidopsis functional genomics: transcript profiling and reverse genetics applications. Genome Res 14:2176–2189
Hirai MY, Yano M, Goodenowe DB, Kanaya S, Kimura T, Awazuhara M, Arita M, Fujiwara T, Saito K (2004) Integration of transcriptomics and metabolomics for understanding of global responses to nutritional stresses in Arabidopsis thaliana. Proc Natl Acad Sci USA 101:10205–10210
Hirai MY, Sugiyama K, Sawada Y, Tohge T, Obayashi T, Suzuki A, Araki R, Sakurai N, Suzuki H, Aoki K, Goda H, Nishizawa OI, Shibata D, Saito K (2007) Omics-based identification of Arabidopsis Myb transcription factors regulating aliphatic glucosinolate biosynthesis. Proc Natl Acad Sci USA 104:6478–6483
Hou XL, Wu P, Jiao FC, Jia QK, Chen HM, Yu J, Song XW, Yi KK (2005) Regulation of the expression of OsIPS1 and OsIPS2 in rice via systemic and local Pi signalling and hormones. Plant Cell Environ 28:353–364
Jung JY, Shin R, Schachtman DP (2009) Ethylene mediates response and tolerance to potassium deprivation in Arabidopsis. Plant Cell 21:607–621
Kant S, Peng M, Rothstein SJ (2011) Genetic regulation by NLA and microRNA827 for maintaining nitrate-dependent phosphate homeostasis in Arabidopsis. PLoS Genet 7:e1002021
Kawashima CG, Yoshimoto N, Maruyama-Nakashita A, Tsuchiya YN, Saito K, Takahashi H, Dalmay T (2009) Sulphur starvation induces the expression of microRNA-395 and one of its target genes but in different cell types. Plant J 57:313–321
Kawashima CG, Matthewman CA, Huang S, Lee BR, Yoshimoto N, Koprivova A, Rubio-Somoza I, Todesco M, Rathjen T, Saito K, Takahashi H, Dalmay T, Kopriva S (2011) Interplay of SLIM1 and miR395 in the regulation of sulfate assimilation in Arabidopsis. Plant J 66:863–876
Kim MJ, Shin R, Schachtman DP (2009) A nuclear factor regulates abscisic acid responses in Arabidopsis. Plant Physiol 151:1433–1445
Krouk G, Crawford NM, Coruzzi GM, Tsay YF (2010a) Nitrate signaling: adaptation to fluctuating environments. Curr Opin Plant Biol 13:266–273
Krouk G, Mirowski P, LeCun Y, Shasha DE, Coruzzi GM (2010b) Predictive network modeling of the high-resolution dynamic plant transcriptome in response to nitrate. Genome Biol 11:R123
Leigh RA, Jones RGW (1984) A hypothesis relating critical potassium concentrations for growth to the distribution and functions of this ion in the plant-cell. New Phytol 97:1–13
Leustek T, Saito K (1999) Sulfate transport and assimilation in plants. Plant Physiol 120:637–644
Li L, Kim BG, Cheong YH, Pandey GK, Luan S (2006) A Ca2+ signaling pathway regulates a K+ channel for low-K response in Arabidopsis. Proc Natl Acad Sci USA 103:12625–12630
Lloyd JC, Zakhleniuk OV (2004) Responses of primary and secondary metabolism to sugar accumulation revealed by microarray expression analysis of the Arabidopsis mutant, pho3. J Exp Bot 55:1221–1230
Lopez-Bucio J, Hernandez-Abreu E, Sanchez-Calderon L, Nieto-Jacobo MF, Simpson J, Herrera-Estrella L (2002) Phosphate availability alters architecture and causes changes in hormone sensitivity in the Arabidopsis root system. Plant Physiol 129:244–256
Lopez-Bucio J, Hernandez-Abreu E, Sanchez-Calderon L, Perez-Torres A, Rampey RA, Bartel B, Herrera-Estrella L (2005) An auxin transport independent pathway is involved in phosphate stress-induced root architectural alterations in Arabidopsis. Identification of BIG as a mediator of auxin in pericycle cell activation. Plant Physiol 137:681–691
Maruyama-Nakashita A, Nakamura Y, Yamaya T, Takahashi H (2004a) A novel regulatory pathway of sulfate uptake in Arabidopsis roots: implication of CRE1/WOL/AHK4-mediated cytokinin-dependent regulation. Plant J 38:779–789
Maruyama-Nakashita A, Nakamura Y, Yamaya T, Takahashi H (2004b) Regulation of high-affinity sulphate transporters in plants: towards systematic analysis of sulphur signalling and regulation. J Exp Bot 55:1843–1849
Maruyama-Nakashita A, Nakamura Y, Watanabe-Takahashi A, Inoue E, Yamaya T, Takahashi H (2005) Identification of a novel cis-acting element conferring sulfur deficiency response in Arabidopsis roots. Plant J 42:305–314
Maruyama-Nakashita A, Nakamura Y, Tohge T, Saito K, Takahashi H (2006) Arabidopsis SLIM1 is a central transcriptional regulator of plant sulfur response and metabolism. Plant Cell 18:3235–3251
Miyake K, Ito T, Senda M, Ishikawa R, Harada T, Niizeki M, Akada S (2003) Isolation of a subfamily of genes for R2R3-MYB transcription factors showing up-regulated expression under nitrogen nutrient-limited conditions. Plant Mol Biol 53:237–245
Nikiforova VJ, Gakiere B, Kempa S, Adamik M, Willmitzer L, Hesse H, Hoefgen R (2004) Towards dissecting nutrient metabolism in plants: a systems biology case study on sulphur metabolism. J Exp Bot 55:1861–1870
Nikiforova VJ, Daub CO, Hesse H, Willmitzer L, Hoefgen R (2005) Integrative gene-metabolite network with implemented causality deciphers informational fluxes of sulphur stress response. J Exp Bot 56:1887–1896
Obertello M, Krouk G, Katari MS, Runko SJ, Coruzzi GM (2010) Modeling the global effect of the basic-leucine zipper transcription factor 1 (bZIP1) on nitrogen and light regulation in Arabidopsis. BMC Syst Biol 4:111
O’Neill EM, Kaffman A, Jolly ER, O’Shea EK (1996) Regulation of PHO4 nuclear localization by the PHO80–PHO85 cyclin-CDK complex. Science 271:209–212
Pant BD, Musialak-Lange M, Nuc P, May P, Buhtz A, Kehr J, Walther D, Scheible WR (2009) Identification of nutrient-responsive Arabidopsis and rapeseed microRNAs by comprehensive real-time polymerase chain reaction profiling and small RNA sequencing. Plant Physiol 150:1541–1555
Peng M, Bi YM, Zhu T, Rothstein SJ (2007a) Genome-wide analysis of Arabidopsis responsive transcriptome to nitrogen limitation and its regulation by the ubiquitin ligase gene NLA. Plant Mol Biol 65:775–797
Peng M, Hannam C, Gu H, Bi YM, Rothstein SJ (2007b) A mutation in NLA, which encodes a RING-type ubiquitin ligase, disrupts the adaptability of Arabidopsis to nitrogen limitation. Plant J 50:320–337
Peng M, Hudson D, Schofield A, Tsao R, Yang R, Gu H, Bi YM, Rothstein SJ (2008) Adaptation of Arabidopsis to nitrogen limitation involves induction of anthocyanin synthesis which is controlled by the NLA gene. J Exp Bot 59:2933–2944
Perez-Torres CA, Lopez-Bucio J, Cruz-Ramirez A, Ibarra-Laclette E, Dharmasiri S, Estelle M, Herrera-Estrella L (2008) Phosphate availability alters lateral root development in Arabidopsis by modulating auxin sensitivity via a mechanism involving the TIR1 auxin receptor. Plant Cell 20:3258–3272
Perez-Torres CA, Lopez Bucio J, Herrera Estrella L (2009) Low phosphate signaling induces changes in cell cycle gene expression by increasing auxin sensitivity in the Arabidopsis root system. Plant Signal Behav 4:781–783
Rouached H, Secco D, Arpat B, Poirier Y (2011) The transcription factor PHR1 plays a key role in the regulation of sulfate shoot-to-root flux upon phosphate starvation in Arabidopsis. BMC Plant Biol 11:19
Rubio V, Linhares F, Solano R, Martin AC, Iglesias J, Leyva A, Paz-Ares J (2001) A conserved MYB transcription factor involved in phosphate starvation signaling both in vascular plants and in unicellular algae. Genes Dev 15:2122–2133
Rubio V, Bustos R, Irigoyen ML, Cardona-Lopez X, Rojas-Triana M, Paz-Ares J (2009) Plant hormones and nutrient signaling. Plant Mol Biol 69:361–373
Saito K (2000) Regulation of sulfate transport and synthesis of sulfur-containing amino acids. Curr Opin Plant Biol 3:188–195
Saito K, Hirai MY, Yonekura-Sakakibara K (2008) Decoding genes with coexpression networks and metabolomics—‘majority report by precogs’. Trends Plant Sci 13:36–43
Schachtman DP, Shin R (2007) Nutrient sensing and signaling: NPKS. Annu Rev Plant Biol 58:47–69
Schachtman DP, Reid RJ, Ayling SM (1998) Phosphorus uptake by plants: from soil to cell. Plant Physiol 116:447–453
Scheible WR, Morcuende R, Czechowski T, Fritz C, Osuna D, Palacios-Rojas N, Schindelasch D, Thimm O, Udvardi MK, Stitt M (2004) Genome-wide reprogramming of primary and secondary metabolism, protein synthesis, cellular growth processes, and the regulatory infrastructure of Arabidopsis in response to nitrogen. Plant Physiol 136:2483–2499
Shin R, Schachtman DP (2004) Hydrogen peroxide mediates plant root cell response to nutrient deprivation. Proc Natl Acad Sci USA 101:8827–8832
Shin R, Berg RH, Schachtman DP (2005) Reactive oxygen species and root hairs in Arabidopsis root response to nitrogen, phosphorus and potassium deficiency. Plant Cell Physiol 46:1350–1357
Shin R, Burch AY, Huppert KA, Tiwari SB, Murphy AS, Guilfoyle TJ, Schachtman DP (2007) The Arabidopsis transcription factor MYB77 modulates auxin signal transduction. Plant Cell 19:2440–2453
Takahashi H, Sasakura N, Noji M, Saito K (1996) Isolation and characterization of a cDNA encoding a sulfate transporter from Arabidopsis thaliana. FEBS Lett 392:95–99
Takahashi H, Yamazaki M, Sasakura N, Watanabe A, Leustek T, Engler JA, Engler G, Van Montagu M, Saito K (1997) Regulation of sulfur assimilation in higher plants: a sulfate transporter induced in sulfate-starved roots plays a central role in Arabidopsis thaliana. Proc Natl Acad Sci USA 94:11102–11107
Takei K, Takahashi T, Sugiyama T, Yamaya T, Sakakibara H (2002) Multiple routes communicating nitrogen availability from roots to shoots: a signal transduction pathway mediated by cytokinin. J Exp Bot 53:971–977
Todd CD, Zeng P, Huete AM, Hoyos ME, Polacco JC (2004) Transcripts of MYB-like genes respond to phosphorous and nitrogen deprivation in Arabidopsis. Planta 219:1003–1009
Tohge T, Nishiyama Y, Hirai MY, Yano M, Nakajima J, Awazuhara M, Inoue E, Takahashi H, Goodenowe DB, Kitayama M, Noji M, Yamazaki M, Saito K (2005) Functional genomics by integrated analysis of metabolome and transcriptome of Arabidopsis plants over-expressing an MYB transcription factor. Plant J 42:218–235
Wang YH, Garvin DF, Kochian LV (2002) Rapid induction of regulatory and transporter genes in response to phosphorus, potassium, and iron deficiencies in tomato roots. Evidence for cross talk and root/rhizosphere-mediated signals. Plant Physiol 130:1361–1370
Yang Y, Yu X, Song L, An C (2011) ABI4 activates DGAT1 expression in Arabidopsis seedlings during nitrogen deficiency. Plant Physiol 156:873–883
Yi K, Wu Z, Zhou J, Du L, Guo L, Wu Y, Wu P (2005) OsPTF1, a novel transcription factor involved in tolerance to phosphate starvation in rice. Plant Physiol 138:2087–2096
Acknowledgement
This work was supported by the RIKEN President’s Discretionary Fund.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Shin, R. Transcriptional Regulatory Components Responding to Macronutrient Limitation. J. Plant Biol. 54, 286–293 (2011). https://doi.org/10.1007/s12374-011-9174-7
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12374-011-9174-7