Abstract
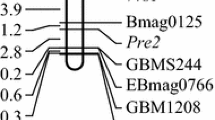
Purple apiculus is one of the important agronomic traits of rice. Single-segment substitution line (SSSL) W23-07-6-02-14 in the genetic background of an elite rice variety Huajingxian74 (HJX74) with the substituted interval of RM225-RM217-RM253 on the chromosome 6 was found to have purple apiculus (Pa). To map the gene governing Pa, W23-07-6-02-14 was crossed with the recipient HJX74 to develop an F2 secondary segregation population. The ratio of purple apiculus to green apiculus showed a good fit to 3:1 ratio, indicating that Pa was controlled by a major dominant gene. The gene locus for Pa was tentatively designated as Pa-6. Using 430 individuals from the F2 segregation population, the Pa-6 locus was mapped between two SSR markers RM19556 and RM19561 with genetic distances of 0.2 and 0.3 cM, respectively. For fine mapping of the Pa-6 gene, a large F2:3 segregation population of 3890 individuals was developed from F2 heterzygous plants in the RM19556-RM19561 region. Recombinant analyses further mapped the Pa-6 gene locus to an interval of 41.7-kb bounded L02 and RM19561. Sequence analysis of this 41.7-kb region revealed that it contains eleven open reading frames (ORFs), of which, ORF5 is classified as the one that is associated with the C (chromogen for anthocyanin) gene, it was presumed to be the candidate gene for Pa. This result provided a foundation of map-based cloning and function analysis of the Pa-6 gene.
Similar content being viewed by others
References
Cone KC, Cocciolone SM, Burr FA, Burr B (1993) Maize anthocyanin regulatory gene p1 is a duplicate of c1 that functions in the plant. The Plant Cell 5:1795–1805
Eshed Y, Zamir D (1995) An introgression line population of Lycopersicon pennellii in the cultivated tomato enables the identification and fine mapping of yield-associated QTL. Genetics 141:1147–1162
Fan FJ, Fan YY, Du JH, Zhuang JY (2008) Fine mapping of C (Chromogen for Anthocyanin) gene in rice. Rice Science 15:1–6
He FH, Xi ZY, Zeng RZ, Tulukdar A, Zhang GQ (2005a) Mapping of heading date QTLs in rice (Oryza sativa L.) using single segment substitution lines, Scientia Agricultura Sinica 38:1505–1513 (In Chinese)
Kinoshita T (1984) Current linkage maps. Rice Genet Newslett 1:16–27
Kishimoto N, Shimosaka E, Matsuura S, Saito A (1992) A current RFLP linkage map of rice: Alignment of the molecular map with the classical map. Rice Genet Newslett 9:118–123
Kosambi, DD (1944) The estimation of map distances from recombination values. Ann Eugen 12:172–175
Lander ES, Gree P (1987) Construction of multilocus genetic linkage maps in human. Proc Natl Acad Sci USA 84:2363–2367
Li WT, Zeng RZ, Zhang ZM, Zhang GQ (2002) Mapping of S-b locus for F1 pollen sterility in cultivated rice using PCR based markers. Acta Bot Sin 44:463–467
Li Z, Pinson SRM, Stansel JW, Park WD (1995) Identification of quantitative trait loci (QTLs) for heading date and plant height in cultivated rice (Oryza sativa L). Theor Appl Genet 91:374–381
Li ZK, Luo LJ, Mei HW, Wang DL, Shu QY, Tabien R, Zhong DB, Ying CS, Stansel JW, Khush GS, Paterson AH (2001) Overdominant epistatic loci are the primary genetic basis of inbreeding depression and heterosis in rice: I. Biomass and grain yield. Genetics 158:1737–1753
Liu GF, Zhang ZM, Zhu HT, Zhao FM, Ding XH, Zeng RZ, Li WT, Zhang GQ (2008) Detection of QTLs with additive effects and additive-by-environment interaction effects on panicle number in rice (Oryza sativa L.) with single segment substitution lines. Theor Appl Genet 116:923–931
Maekawa M, Kita F (1987) Genic analysis for weakness induced with gamma-ray treatment and mechanisms for the occurrence of triploid plants of rice. Jpn J Breed 37(Suppl. 2):308–309 (In Japanese)
McCouch SR, Kochert G, Yu ZH, Wang ZY, Khush GS, Coffman WR, Tanksley SD (1988) Molecular mapping of rice chromosomes. Theor Appl Genet 76:815–829
Mikami I, Takahashi A, Khin-thidar A, Sano Y (2000) A candidate for C (Chromogen for anthocyanin) gene. Rice Genet Newslett 17:54–56
Mori KI, Takahashi ME (1981) Differentiation of multiple alleles for anthocyanin color character of apiculus in indica rice varietiesgenetical studies on rice plants, LXXXI. Japan J Breed 31: 226–238
Nagao S, Takahashi T (1963) Trial construction of twelve linkage groups in Japanese rice. Genetical studies of rice plant, XXVII, J. Fac Agric Hokkaido Univ 53:72–230
Reddy AR (1996) Genetic and molecular analysis of the anthocyanin pigmentation pathway in rice. In: Khush G S. Rice Genetics III. Manila, Philippines: IRRI:341–352
Reddy AR, Scheffler B, Srivastava MN, Kumar A, Sathyanarayan P V, Nair S, Mohan M (1995) Chalcone synthase (CHS) gene in rice (Oryza sativa L): expression and molecular mapping. Plant Genome III, San Diego, California, USA. P121
Reddy VS, Dash S, Reddy AR (1995) Anthocyanin pathway in rice (Oryza sativa L): identification of a mutant showing dominant inhibition of anthocyanins in leaf and accumulation of proanthocyanidins in pericarp. Theor Appl Genet 91:301–312
Saitoh, K, Onishi K. Mikami I, Thidar K, Sano Y (2004) Alleic diversification at the C (OsC1) locus of wild and cultivated rice: nucleotide changes associated with phenotypes. Genetics 168:997–1007
Takahashi ME (1982) Gene analysis and its related problems. J. Fac. Agr. Hokkaido Univ 61:91–142
Wang J, Liu KD, Xu CG, Li XH, Zhang QF (1998) The high level of wide-compatibility of variety “Dular” has a complex genetic basis. Theor Appl Genet 97:407–412
Wang WY, Ding HF, Li GX, Jiang MS, Li RF, Liu X, Zhang Y, Yao FY (2009) Delimitation of the PSH1(t) gene for rice purple leaf sheath to a 23.5-kb DNA fragment. Genome 52:268–274
Xi ZY, He FH, Zeng RZ, Zhang ZM, Ding XH, Li WT, Zhang GQ (2006) Development of a wide population of chromosome single-segment substitution lines in the genetic background of an elite cultivar of rice (Oryza sativa L.) Genome 49:476–484
Yue B, Cui KH, Yu SB, Xue WY, Luo LJ, Xing YZ (2006) Molecular marker-assisted dissection of quantitative trait loci for seven morphological traits in rice (Oryza sativa L.). Euphytica 150: 131–139
Zhang GQ, Zeng RZ, Zhang ZM, Ding XH, Li WT, Liu GM, He FH, Tulukdar A, Huang CF, Xi ZY, Qin LJ, Shi JQ, Zhao FM, Feng MJ, Shan ZL, Chen L, Guo XQ, Zhu HT, Lu YG (2004) The construction of a library of single segment substitution lines in rice (Oryza sativa L.) Rice Genetics Newsletter 21:85–87
Zhao FM, Liu GF, Zhu HT, Ding XH, Zeng RZ, Zhang ZM, Li WT, Zhang GQ (2008) Unconditional and conditional QTL mapping for tiller number at various stages by using single segment substitution lines in rice (Oryza sativa L.). Agricultural Sciences in China 7:257–265
Zheng KL, Huang N, Bennett J, Khush GS (1995) PCR-based marker-assisted selection in rice breeding. In IRRI Discussion Paper Series. No. 12. International Rice Research Institute, Manila, Philippines
Author information
Authors and Affiliations
Corresponding author
Additional information
These authors contributed equally to this work
Rights and permissions
About this article
Cite this article
Liu, X., Sun, X., Wang, W. et al. Fine mapping of Pa-6 gene for purple apiculus in rice. J. Plant Biol. 55, 218–225 (2012). https://doi.org/10.1007/s12374-011-0276-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12374-011-0276-z