Abstract
Background
Deletions involving the long arm of chromosome 6 have been reported to occur in breast cancer, but little is known about the clinical relevance of this alteration.
Methods
We made use of a pre-existing tissue microarray with 2197 breast cancers and employed a 6q15/centromere 6 dual-labeling probe for fluorescence in situ (FISH) analysis
Results
Heterozygous 6q15 deletions were found in 202 (18%) of 1099 interpretable cancers, including 19% of 804 cancers of no special type (NST), 3% of 29 lobular cancers, 7% of 41 cribriform cancers, and 28% of 18 cancers with papillary features. Homozygous deletions were not detected. In the largest subset of NST tumors, 6q15 deletions were significantly linked to advanced tumor stage and high grade (p < 0.0001 each). 6q deletions were also associated with estrogen receptor negativity (p = 0.0182), high Ki67 proliferation index (p < 0.0001), amplifications of HER2 (p = 0.0159), CCND1 (p = 0.0069), and cMYC (p = 0.0411), as well as deletions of PTEN (p = 0.0003), 8p21 (p < 0.0001), and 9p21 (p = 0.0179). However, 6q15 deletion was unrelated to patient survival in all cancers, in NST cancers, or in subsets of cancers defined by the presence or absence of lymph-node metastases.
Conclusion
Our data demonstrate that 6q deletion is a frequent event in breast cancer that is statistically linked to unfavorable tumor phenotype and features of genomic instability. The absence of any prognostic impact argues against a clinical applicability of 6q15 deletion testing in breast cancer patients.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Breast cancer is the most common malignancy detected in women [1]. Surgical removal of the cancer represents the standard of care. Whether or not adjuvant systemic therapy is given depends on the perceived aggressiveness of the removed cancer. Currently established prognostic parameters mainly include histological grade, tumor size, presence of lymph-node metastasis, tumor cell proliferation (Ki67-labeling index; Ki67 LI), as well as hormonal receptor and HER2 status [2]. Additional molecular parameters are analyzed in many patients [3,4,5]. Commercial molecular classifiers are based on multiplexed analyses of the RNAs of 21–70 gene products [6,7,8]. These purely RNA-based tests share the disadvantage that gradual changes of each parameter must be measured, and that these measurements are strongly dependent on tumor cell purity. As next-generation sequencing (NGS) is getting less expensive, it is expected that alternative and potentially better prognostic tests will be increasingly based on DNA analyses including a global assessment of structural rearrangements and gene mutations. NGS tests can analyze biomarkers with yes/no answers such as presence or absence of individual mutations or deletions. In other tumor types, especially in prostate cancer—another important hormone dependent cancer—various chromosomal deletions have been shown to have substantial prognostic relevance [9,10,11,12]. One of these is deletion of 6q12-q21, which is also commonly found in breast cancer. Studies using classical comparative genomic hybridization in 16–34 patients [13, 14], array-based copy-number screening assays in 28 patients [15], or loss of heterozygosity (LOH) analysis 42–83 patients [14, 16,17,18] reported 6q deletions in 6–50% of breast cancers. Some of these studies have described an association of 6q deletions with unfavorable tumor phenotype [14, 19].
To better understand the clinical relevance of 6q deletions in breast cancer, we utilized a pre-existing breast cancer tissue microarray (TMA) containing more than 2000 cancers. Our data show that 6q deletion is frequent but unrelated to patient prognosis in breast cancer.
Materials and methods
Breast cancer tissue microarray (TMA)
A pre-existing tissue microarray (TMA) was used for this study [20]. The TMA contained 2197 human breast cancer tissue punches (diameter 0.6 mm) from paraffin-embedded tissue specimens fixed in 4% neutral buffered formalin. The donor blocks used for TMA construction were collected from the archives of the Institute of Pathology of the University Hospital Basel, the Institute for Clinical Pathology in Basel, and the Triemli Hospital in Zurich. Tumors were collected consecutively, and all slides from the tumors were reviewed by specialized pathologists to define the histologic grade according to Elston and Ellis [21] and the histologic tumor type. The use of the specimens and data for research purposes were approved by the Ethics Committee of the Basel University Hospital. Survival data were either obtained from the cancer registry of Basel or collected from the patients attending physicians. The median patient’s age was 63 (range 26–101) years. Raw survival data were available from 1982 patients (713 patients with and 1508 without event). The mean follow-up time was 63 months (range 1–176 months). Tumor size and nodal status were obtained from the primary pathology reports. Four micrometer sections of the TMA blocks were transferred to an adhesive-coated slide system (Instrumedics Inc., Hackensack, New Jersey) for FISH analysis. Molecular data used in this study were available from previously published studies. These included data obtained by FISH for amplification of HER2 [20, 22], CCND1 [20], MDM2 [20], and cMYC [20, 22] as well as for deletions of PTEN [23], 8p21 [24], and 9p21 [25] and data obtained by IHC for estrogen receptor (ER) and progesterone receptor (PR) expression as well as Ki67-labeling index (Ki67 LI) [20, 26]. Molecular subtypes (Luminal A, B, HER2, basal cell type) were defined according to the St. Gallen (2011) criteria [27].
Fluorescence in situ hybridization
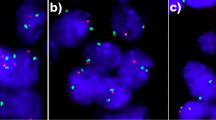
Four micrometer TMA sections were used for FISH. For proteolytic slide pretreatment, a commercial kit was used (paraffin pretreatment reagent kit; Abbott, Wiesbaden, Germany). TMA sections were deparaffinized, air-dried, and dehydrated in 70%, 85%, and 100% ethanol, followed by denaturation for 5 min at 74 °C in 70% formamide 2 × SSC solution. The FISH probe set consisted of a spectrum-green labeled 6q15 (MAP3K7) probe (made from a mixture of BAC RP3-470J08 and BAC RP11-501P02), and a spectrum-orange labeled commercial centromere 6 probe (#06J36-06; Abbott, Wiesbaden, Germany) as a reference. Hybridization was performed overnight at 37 °C in a humidified chamber. Slides were subsequently washed and counterstained with 0.2 µmol/L 4’-6-diamidino-2-phenylindole in antifade solution. Stained slides were manually interpreted with an epifluorescence microscope, and the predominant FISH signal numbers were recorded in each tissue spot. The presence of fewer 6q15 signals than centromere 6 probe signals in at least 60% tumor nuclei was considered a heterozygous deletion. These thresholds were based on our previous study analyzing PTEN deletions on a prostate cancer TMA where our approach resulted in a 100% concordance with array comparative genomic hybridization (CGH) data [12]. Complete absence of 6q15 signals in all tumor cells, but presence of centromere 6 and 6q15 signals in adjacent normal cells, was considered a homozygous deletion. Tissue spots lacking any detectable 6q15 signals in all cells (tumor and normal cells or tumor cells only but no normal cells present) were excluded from analysis because of a lack of an internal control for successful hybridization of the 6q15 probe. Representative images of 6q15 FISH results are shown in Fig. 1.
Statistics
Statistical calculations were performed with JMP 14 software (SAS Institute Inc., NC, USA). Contingency table analysis and Chi-square test were used to study the relationship between FISH results and clinicopathological variables. Kaplan–Meier plots were used to estimate overall survival and the statistical significance was determined by the log-rank test. The log-rank test was applied to test the significance of differences between stratified survival functions.
Results
Technical issues
A total of 1099 (50%) of arrayed cancer samples were analyzable by FISH. Reasons for non-informative results included non-interpretable FISH signals (589, 26%), lack of tumor cells in the tissue spot (224, 10%), or absence of tissue spot on the TMA section (309, 14%).
6q15 deletion and breast cancer phenotype
Heterozygous 6q15 deletions were found in 202 (18%) interpretable breast cancers. Representative images from cancers with and without 6q15 deletions are given in Fig. 1a,b. Homozygous 6q15 deletion was not observed. 6q15 deletions were found in 19% of 804 NST cancers, 3% of 29 tubular cancers (p = 0.0076 vs NST), 7% of 41 cribriform cancers (p = 0.0338 vs NST), and 28% of 18 cancers with papillary features (p = 0.5508 vs NST). If all cancers were jointly analyzed, deletion of 6q15 was significantly linked to advance tumor stage (p = 0.0315) and high histopathological grade (p < 0.0001). These associations also held true for BRE grade in the largest subset of NST cancers (p < 0.0001). In addition, 6q15 deletions were significantly linked to the subset of estrogen receptor (ER) negative breast cancers: deletion was found in 24% of ER negative but only in 17% of ER-positive breast cancers (p = 0.0182) and to the molecular subtypes of HER2-positive and basal cell type cancers (p = 0.0062). 6q15 deletion was unrelated to the presence of lymph-node metastases and progesterone receptor status. All results are summarized in Table 1.
6q15 deletion and tumor cell proliferation
Data on tumor cell proliferation, as determined by immunohistochemical analysis of the Ki67 antigen, were available from a previous study using the same TMA [20]. Deletion of 6q15 was tightly linked to a high Ki67 LI if all cancers were jointly analyzed (p < 0.0001). This association was not independent of histological grade. All results are summarized in Table 2.
Prognostic significance of 6q15 deletion
Data on raw survival were available from 1097 cancers with interpretable 6q15 FISH results. The presence of 6q15 deletion was largely unrelated to shortened overall survival if all cancers were jointly analyzed (p = 0.6709, Fig. 2a), as well as in the subsets of cancers of No Special Type (NST, p = 0.3317, Fig. 2b), in the subset of NST cancers with nodal metastases (p = 0.5635, Fig. 2c), and in the subset of cancers with or without nodal metastases (p = 0.5844 for pN positive and p = 0.9741 for pN negative; Fig. 2d-e).
6q15 deletion and other genomic alterations in breast cancer
HER2, CCND1, MYC, and MDM2 amplification, as well as PTEN, 8p21, and 9p21 deletion results were available from our previous studies. In total, FISH results on both 6q15 deletions and alterations of HER2, CCND1, MYC, MDM2, PTEN, 8p21, and 9p21 were available in subsets of 921 (HER2), 1007 (CCND1), 699 (MYC), 1022 (MDM2), 980 (PTEN), 986 (8p21), and 902 (9p21) cancers. Deletions of 6q15 were significantly linked to most of all alterations (p ≤ 0.04). For example, 6q15 deletion was found in 25% of 173 HER2-amplified cancers but only in 17% of 748 cancers with normal HER2 copy-number status (p = 0.0159), as well as in 26% of 185 PTEN deleted cancers but only in 17% of 795 cancers with normal PTEN status (p = 0.0030). No significant association was found between 6q15 deletion and MDM2 amplification (p = 0.1750). All results are summarized in Fig. 3.
Discussion
The analysis of more than 1000 breast cancers using an FISH probe directed against a DNA sequence at 6q15 identified a heterozygous deletion in 18% of tumors. This finding is consistent with data from the ICGC/TCGA database describing 6q15 deletion in 26% of 2051 sequenced breast cancers from the METABRIC cohort [28]. Earlier published studies had reported 6q deletion frequencies between 6 and 50% in cohorts of 16–83 patients [13,14,15,16,17,18,19] and in 8 of 10 breast cancer cell lines [29]. A variety of methods had been applied in these studies including classical and array CGH, LOH analysis, and FISH. We are confident that our findings reflect the true frequency of 6q15 deletion in breast cancer, because fluorescence in situ hybridization (FISH) represents the gold standard for gene copy-number analysis. FISH is independent of the purity of cancer tissue and chromosomal aberrations such as polysomy. Deletions can be analyzed on a cell-by-cell basis, and abnormalities can be detected in a few cells or even a single cell. In our study, 6q15 deletion was defined as “fewer 6q15 signals than centromere 6 signals in at least 60% of all tumor cells”. These stringent criteria resulted in a 100% concordance of results found by FISH and comparative genomic hybridization in a previous PTEN study of our group in prostate cancer [12]. Our cut-off of 60% deleted cells is also supported by the observation that virtually all deleted cases had fewer gene signals than centromere signals in > 80% of cells and undeleted cases had usually fewer than 10% cells with fewer 6q15 signals than centromere signals.
The comparison of 6q15 deletions with histopathological tumor features revealed statistically significant associations. This was true for aggressive molecular subtypes including the HER2 and basal cell type and especially for the BRE grade. This is not surprising as a high BRE grade is characterized by a particular high degree of nuclear atypia, which—in turn—is often related to a high frequency of genomic alterations [30,31,32]. Accordingly, the rate of 6q15 deletions was particularly low in cancer subtypes characterized by low nuclear atypia such as tubular or lobular carcinoma and particularly high in medullary carcinoma, a tumor characterized by substantial nuclear atypia. The assumption that 6q15 deletions accumulate in tumors with an increased level of genomic instability which fits well with the significant associations found between 6q15 deletions and all other previously analyzed genomic aberrations such as amplifications of MYC; HER2, MDM2, and CCND1, as well as deletions of 8p, 9p21 and PTEN [20, 22,23,24,25]. The relationship of 6q15 deletions with all these aberrations was highly similar. All genomic changes occurred between 1.4 and 1.8 more often in 6q15 deleted than in 6q15 undeleted carcinomas. It appears thus more likely that these associations are caused by a general phenomenon such as “genomic instability” than by specific interactions between associated pathways. In an earlier study analyzing gene amplifications, we had already found that tumors carrying one amplification are significantly more prone to develop additional amplifications [33].
The 6q gene(s) driving cancer progression through inactivation has not been clearly identified. Copy-number data derived from the ICGC/TCGA database (www.cbioportal.org) [34] do not suggest a clear-cut minimal commonly deleted region in breast cancer, although highest frequencies (26%) are found in the interval between 85 Mb (6q14) and 100 Mb (6q21). An FISH probe for MAP3K7 had been selected for this study because of its location in the center of the 6q15 deletion and the known tumor suppressive function of MAP3K7 [35]. Other 6q15 genes with potential tumor suppressive functions for example include EEF1A1 [36], ZNF292 [37], SNORD50A [38], PRDM1 [39], CCNC [40], FOXO3 [41], WISP3 [42], and FRK [43]. It is of note, however, that inactivation of the second allele by homozygous deletion or inactivating mutation is virtually not existent. In the METABRIC [28] dataset, EEF1A1 or FOXO3 were the only genes for which homozygous deletions could be seen in 1.6% of more than 1,000 tumors. A classical tumor suppressive role of FOXO3 in a very small subset of breast cancers is further supported by recurrent mutations (n = 38, 1.4%), almost half of them being associated with deletions of the second allele. For all other 6q15 candidate genes, neither homozygous deletions nor recurrent mutations were described. 6q15 deletions—as other large genomic deletions—may thus exert their tumor promoting role through a reduced function of multiple genes within the deletion. Of note, the complete absence of large homozygous MAP3K7 deletions argues for one or several essential genes in the 6q15 area for which complete inactivated is not consistent with cell survival.
Irrespective of which gene(s) are affected by 6q15 deletions, our data do not suggest a substantial impact of a dysfunction of these for the clinical course of affected patients. The complete lack of differences in patient outcome between 6q15 deleted and undeleted cancers would even be consistent with 6q15 deletions representing an irrelevant “passenger” lesion” in breast cancer. However, given the prominent role of 6q15 deletions in various other cancer types, including a clear-cut prognostic impact in prostate cancer [44, 45], we would not anticipate this deletion to be meaningless. There are examples of critical molecular events for cancer development, such as TMPRSS2–ERG fusions, the most frequent molecular alteration in prostate cancer occurring in about 50% of cases, which are completely unrelated to disease outcome [46]. Our data on “only” 1099 successfully analyzed cancers do not exclude a clinically relevant role of 6q15 alterations in a morphologically, molecularly, or clinically (treatment) defined subgroup of cancers.
In conclusion, these data identify 6q15 deletions as a frequent event in breast cancer. Despite statistically significant associations with important histological and molecular features, 6q15 deletions are largely unrelated to patient outcome. 6q15 deletion analysis does not appear to have potential clinical utility.
References
Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global Cancer Statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71:209–49.
Duffy MJ, Harbeck N, Nap M, Molina R, Nicolini A, Senkus E, et al. Clinical use of biomarkers in breast cancer: updated guidelines from the European Group on Tumor Markers (EGTM). Eur J Cancer. 2017;75:284–98.
Giuliano AE, Hunt KK, Ballman KV, Beitsch PD, Whitworth PW, Blumencranz PW, et al. Axillary dissection vs no axillary dissection in women with invasive breast cancer and sentinel node metastasis: a randomized clinical trial. JAMA. 2011;305:569–75.
McVeigh TP, Hughes LM, Miller N, Sheehan M, Keane M, Sweeney KJ, et al. The impact of Oncotype DX testing on breast cancer management and chemotherapy prescribing patterns in a tertiary referral centre. Eur J Cancer. 2014;50:2763–70.
Naoi Y, Noguchi S. Multi-gene classifiers for prediction of recurrence in breast cancer patients. Breast Cancer. 2016;23:12–8.
Hornberger J, Cosler LE, Lyman GH. Economic analysis of targeting chemotherapy using a 21-gene RT-PCR assay in lymph-node-negative, estrogen-receptor-positive, early-stage breast cancer. Am J Manag Care. 2005;11:313–24.
Cobleigh MA, Tabesh B, Bitterman P, Baker J, Cronin M, Liu ML, et al. Tumor gene expression and prognosis in breast cancer patients with 10 or more positive lymph nodes. Clin Cancer Res. 2005;11:8623–31.
van 't Veer LJ, Dai H, van de Vijver MJ, He YD, Hart AA, Mao M, et al. Gene expression profiling predicts clinical outcome of breast cancer. Nature. 2002; 415: 530–6.
Krohn A, Seidel A, Burkhardt L, Bachmann F, Mader M, Grupp K, et al. Recurrent deletion of 3p13 targets multiple tumour suppressor genes and defines a distinct subgroup of aggressive ERG fusion-positive prostate cancers. J Pathol. 2013;231:130–41.
El Gammal AT, Bruchmann M, Zustin J, Isbarn H, Hellwinkel OJ, Kollermann J, et al. Chromosome 8p deletions and 8q gains are associated with tumor progression and poor prognosis in prostate cancer. Clin Cancer Res. 2010;16:56–64.
Kluth M, Harasimowicz S, Burkhardt L, Grupp K, Krohn A, Prien K, et al. Clinical significance of different types of p53 gene alteration in surgically treated prostate cancer. Int J Cancer. 2014;135:1369–80.
Krohn A, Diedler T, Burkhardt L, Mayer PS, De Silva C, Meyer-Kornblum M, et al. Genomic deletion of PTEN is associated with tumor progression and early PSA recurrence in ERG fusion-positive and fusion-negative prostate cancer. Am J Pathol. 2012;181:401–12.
Nishizaki T, DeVries S, Chew K, Goodson WH 3rd, Ljung BM, Thor A, et al. Genetic alterations in primary breast cancers and their metastases: direct comparison using modified comparative genomic hybridization. Genes Chromosomes Cancer. 1997;19:267–72.
Rodriguez C, Causse A, Ursule E, Theillet C. At least five regions of imbalance on 6q in breast tumors, combining losses and gains. Genes Chromosomes Cancer. 2000;27:76–84.
Han W, Han MR, Kang JJ, Bae JY, Lee JH, Bae YJ, et al. Genomic alterations identified by array comparative genomic hybridization as prognostic markers in tamoxifen-treated estrogen receptor-positive breast cancer. BMC Cancer. 2006;6:92.
Orphanos V, McGown G, Hey Y, Boyle JM, Santibanez-Koref M. Proximal 6q, a region showing allele loss in primary breast cancer. Br J Cancer. 1995;71:290–3.
Sheng ZM, Marchetti A, Buttitta F, Champeme MH, Campani D, Bistocchi M, et al. Multiple regions of chromosome 6q affected by loss of heterozygosity in primary human breast carcinomas. Br J Cancer. 1996;73:144–7.
Noviello C, Courjal F, Theillet C. Loss of heterozygosity on the long arm of chromosome 6 in breast cancer: possibly four regions of deletion. Clin Cancer Res. 1996;2:1601–6.
Devilee P, van Vliet M, van Sloun P, Kuipers Dijkshoorn N, Hermans J, Pearson PL, et al. Allelotype of human breast carcinoma: a second major site for loss of heterozygosity is on chromosome 6q. Oncogene. 1991;6:1705–11.
Ruiz C, Seibt S, Al Kuraya K, Siraj AK, Mirlacher M, Schraml P, et al. Tissue microarrays for comparing molecular features with proliferation activity in breast cancer. Int J Cancer. 2006;118:2190–4.
Elston CW, Ellis IO. Pathological prognostic factors in breast cancer. I. The value of histological grade in breast cancer: experience from a large study with long-term follow-up. Histopathology. 2002; 41: 154–61.
Al-Kuraya K, Schraml P, Torhorst J, Tapia C, Zaharieva B, Novotny H, et al. Prognostic relevance of gene amplifications and coamplifications in breast cancer. Cancer Res. 2004;64:8534–40.
Lebok P, Kopperschmidt V, Kluth M, Hube-Magg C, Ozden C, B T, et al. Partial PTEN deletion is linked to poor prognosis in breast cancer. BMC Cancer. 2015; 15: 963.
Lebok P, Mittenzwei A, Kluth M, Ozden C, Taskin B, Hussein K, et al. 8p deletion is strongly linked to poor prognosis in breast cancer. Cancer Biol Ther. 2015;16:1080–7.
Lebok P, Roming M, Kluth M, Koop C, Ozden C, Taskin B, et al. p16 overexpression and 9p21 deletion are linked to unfavorable tumor phenotype in breast cancer. Oncotarget. 2016;7:81322–31.
Al Kuraya K, Simon R, Sauter G. Tissue microarrays for high-throughput molecular pathology. Ann Saudi Med. 2004;24:169–74.
Goldhirsch A, Wood WC, Coates AS, Gelber RD, Thurlimann B, Senn HJ, et al. Strategies for subtypes--dealing with the diversity of breast cancer: highlights of the St. Gallen International Expert Consensus on the Primary Therapy of Early Breast Cancer 2011. Ann Oncol. 2011; 22: 1736–47.
Pereira B, Chin SF, Rueda OM, Vollan HK, Provenzano E, Bardwell HA, et al. The somatic mutation profiles of 2433 breast cancers refines their genomic and transcriptomic landscapes. Nat Commun. 2016;7:11479.
Zhang Y, Matthiesen P, Siebert R, Harder S, Theile M, Scherneck S, et al. Detection of 6q deletions in breast carcinoma cell lines by fluorescence in situ hybridization. Hum Genet. 1998;103:727–9.
Reis-Filho JS, Simpson PT, Gale T, Lakhani SR. The molecular genetics of breast cancer: the contribution of comparative genomic hybridization. Pathol Res Pract. 2005;201:713–25.
Bowtell DD. The genesis and evolution of high-grade serous ovarian cancer. Nat Rev Cancer. 2010;10:803–8.
Ried T, Heselmeyer-Haddad K, Blegen H, Schrock E, Auer G. Genomic changes defining the genesis, progression, and malignancy potential in solid human tumors: a phenotype/genotype correlation. Genes Chromosomes Cancer. 1999;25:195–204.
Choschzick M, Lassen P, Lebeau A, Marx AH, Terracciano L, Heilenkotter U, et al. Amplification of 8q21 in breast cancer is independent of MYC and associated with poor patient outcome. Mod Pathol. 2010;23:603–10.
Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401–4.
Wu M, Shi L, Cimic A, Romero L, Sui G, Lees CJ, et al. Suppression of Tak1 promotes prostate tumorigenesis. Cancer Res. 2012;72:2833–43.
Kim MS, Jeong EG, Chung YJ, Yoo NJ, Lee SH. Absence of somatic mutation of a tumor suppressor gene eukaryotic translation elongation factor 1, epsilon-1 (EEF1E1), in common human cancers. APMIS. 2008;116:832–3.
Lee JH, Song SY, Kim MS, Yoo NJ, Lee SH. Frameshift mutations of a tumor suppressor gene ZNF292 in gastric and colorectal cancers with high microsatellite instability. APMIS. 2016;124:556–60.
Dong XY, Rodriguez C, Guo P, Sun X, Talbot JT, Zhou W, et al. SnoRNA U50 is a candidate tumor-suppressor gene at 6q14.3 with a mutation associated with clinically significant prostate cancer. Hum Mol Genet. 2008; 17: 1031–42.
Kang HB, Lee HR, da Jee J, Shin SH, Nah SS, Yoon SY, et al. PRDM1, a Tumor-Suppressor Gene, is Induced by Genkwadaphnin in Human Colon Cancer SW620 Cells. J Cell Biochem. 2016;117:172–9.
Ohata N, Ito S, Yoshida A, Kunisada T, Numoto K, Jitsumori Y, et al. Highly frequent allelic loss of chromosome 6q16-23 in osteosarcoma: involvement of cyclin C in osteosarcoma. Int J Mol Med. 2006;18:1153–8.
Shin H, Cha HJ, Na K, Lee MJ, Cho JY, Kim CY, et al. O-GlcNAcylation of the tumor suppressor FOXO3 triggers aberrant cancer cell growth. Cancer Res. 2018;78:1214–24.
Tran MN, Kleer CG. Matricellular CCN6 (WISP3) protein: a tumor suppressor for mammary metaplastic carcinomas. J Cell Commun Signal. 2018;12:13–9.
Yim EK, Peng G, Dai H, Hu R, Li K, Lu Y, et al. Rak functions as a tumor suppressor by regulating PTEN protein stability and function. Cancer Cell. 2009;15:304–14.
Liu W, Chang BL, Cramer S, Koty PP, Li T, Sun J, et al. Deletion of a small consensus region at 6q15, including the MAP3K7 gene, is significantly associated with high-grade prostate cancers. Clin Cancer Res. 2007;13:5028–33.
Kluth M, Hesse J, Heinl A, Krohn A, Steurer S, Sirma H, et al. Genomic deletion of MAP3K7 at 6q12-22 is associated with early PSA recurrence in prostate cancer and absence of TMPRSS2:ERG fusions. Mod Pathol. 2013;26:975–83.
Minner S, Enodien M, Sirma H, Luebke AM, Krohn A, Mayer PS, et al. ERG status is unrelated to PSA recurrence in radically operated prostate cancer in the absence of antihormonal therapy. Clin Cancer Res. 2011;17:5878–88.
Acknowledgements
We thank Janett Lütgens, Sünje Seekamp, and Inge Brandt for excellent technical assistance.
Funding
Open Access funding enabled and organized by Projekt DEAL. No funding.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
We certify that there is no actual or potential conflict of interest in relation to this article.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
This article is published under an open access license. Please check the 'Copyright Information' section either on this page or in the PDF for details of this license and what re-use is permitted. If your intended use exceeds what is permitted by the license or if you are unable to locate the licence and re-use information, please contact the Rights and Permissions team.
About this article
Cite this article
Lebok, P., Bönte, H., Kluth, M. et al. 6q deletion is frequent but unrelated to patient prognosis in breast cancer. Breast Cancer 29, 216–223 (2022). https://doi.org/10.1007/s12282-021-01301-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12282-021-01301-5