Abstract
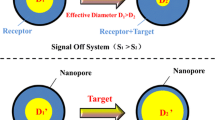
Conventional solid-state nanopore measurements sense all translocating entities, necessitating meticulous analysis to differentiate target biomolecules. To address this, we have established a selective assay with the platform that has shown utility in quantifying several nucleic acid biomarkers. However, limited detection efficiency and intrinsic noise have so far limited assay resolution to 10 nM. Improvements in this value require manipulation of translocation dynamics. Here, we report the effects of NaCl conditions on assay performance. We first investigate symmetric conditions, finding sensitivity increases with salt concentration but selectivity is maximized at 1.0 M NaCl. We then probe asymmetric conditions, showing a remarkable impact on assay sensitivity and selectivity when measurement buffer NaCl concentration in the reservoir with the translocating molecules is low and the opposite reservoir is increased. Using optimum conditions, we demonstrate detection of target biomolecules down to a concentration of 100 pM which is an improvement of 2 orders of magnitude over past results.

Similar content being viewed by others
References
Srinivas, P. R.; Kramer, B. S.; Srivastava, S. Trends in biomarker research for cancer detection. Lancet Oncol. 2001, 2, 698–704.
Qureshi, A.; Gurbuz, Y.; Niazi, J. H. Biosensors for cardiac biomarkers detection: A review. Sens. Actuators B: Chem. 2012, 171-172, 62–76.
Berg, D. Biomarkers for the early detection of Parkinson’s and Alzheimer’s disease. Neurodegener. Dis. 2008, 5, 133–136.
Rysz, J.; Gluba-Brzózka, A.; Franczyk, B.; Jablonowski, Z.; Cialkowska-Rysz, A. Novel biomarkers in the diagnosis of chronic kidney disease and the prediction of its outcome. Int. J. Mol. Sci. 2017, 18, 1702.
Broza, Y. Y.; Zhou, X.; Yuan, M. M.; Qu, D. Y.; Zheng, Y. B.; Vishinkin, R.; Khatib, M.; Wu, W. W.; Haick, H. Disease detection with molecular biomarkers: From chemistry of body fluids to nature-inspired chemical sensors. Chem. Rev. 2019, 119, 11761–11817.
Wishart, D. S.; Bartok, B.; Oler, E.; Liang, K. Y. H.; Budinski, Z.; Berjanskii, M.; Guo, A.; Cao, X.; Wilson, M. MarkerDB: An online database of molecular biomarkers. Nucleic Acids Res. 2021, 49, D1259–D1267.
Erel, O. A new automated colorimetric method for measuring total oxidant status. Clin. Biochem. 2005, 38, 1103–1111.
Liu, J. W.; Lu, Y. Fast colorimetric sensing of adenosine and cocaine based on a general sensor design involving aptamers and nanoparticles. Angew. Chem., Int. Ed. 2005, 45, 90–94.
Liu, R. T.; Ye, X. Y.; Cui, T. H. Recent progress of biomarker detection sensors. Research 2020, 2020, 7949037.
Huang, J.; Du, J.; Cevher, Z.; Ren, Y. H.; Wu, X. H.; Chu, Y. L. Printable and flexible phototransistors based on blend of organic semiconductor and biopolymer. Adv. Funct. Mater. 2017, 27, 1604163.
Yang, Y. J.; Zeng, B.; Li, Y. X.; Liang, H. G.; Yang, Y. B.; Yuan, Q. Construction of MoS2 field effect transistor sensor array for the detection of bladder cancer biomarkers. Sci. China Chem. 2020, 63, 997–1003.
Hawkridge, A. M.; Muddiman, D. C. Mass spectrometry-based biomarker discovery: Toward a global proteome index of individuality. Annu. Rev. Anal. Chem. (Palo Alto Calif) 2009, 2, 265–277.
Li, C.; Yang, Y. C.; Wu, D.; Li, T. Q.; Yin, Y. M.; Li, G. X. Improvement of enzyme-linked immunosorbent assay for the multicolor detection of biomarkers. Chem. Sci. 2016, 7, 3011–3016.
Hajian, R.; Balderston, S.; Tran, T.; deBoer, T.; Etienne, J.; Sandhu, M.; Wauford, N. A.; Chung, J. Y.; Nokes, J.; Athaiya, M.; Paredes, J. et al. Detection of unamplified target genes via CRISPR-Cas9 immobilized on a graphene field-effect transistor. Nat. Biomed. Eng. 2019, 3, 427–437.
Kong, D. R.; Wang, X. J.; Gu, C. J.; Guo, M. Q.; Wang, Y.; Ai, Z. L.; Zhang, S.; Chen, Y. H.; Liu, W. T.; Wu, Y. G. et al. Direct SARS-CoV-2 nucleic acid detection by Y-shaped DNA dual-probe transistor assay. J. Am. Chem. Soc. 2021, 143, 17004–17014.
Thomou, T.; Mori, M. A.; Dreyfuss, J. M.; Konishi, M.; Sakaguchi, M.; Wolfrum, C.; Rao, T. N.; Winnay, J. N.; Garcia-Martin, R.; Grinspoon, S. K. et al. Adipose-derived circulating MiRNAs regulate gene expression in other tissues. Nature 2017, 542, 450–455.
Kanagawa, T. Bias and artifacts in multitemplate polymerase chain reactions (PCR). J. Biosci. Bioeng. 2003, 96, 317–323.
Chen, C. F.; Tan, R. Y.; Wong, L. D.; Fekete, R.; Halsey, J. Quantitation of microRNAs by real-time RT-qPCR. Methods Mol. Biol. 2011, 687, 113–134.
Grunau, C.; Clark, S. J.; Rosenthal, A. Bisulfite genomic sequencing: Systematic investigation of critical experimental parameters. Nucleic Acids Res. 2001, 29, e65.
Yu, M.; Hon, G. C.; Szulwach, K. E.; Song, C. X.; Jin, P.; Ren, B.; He, C. Tet-assisted bisulfite sequencing of 5-hydroxymethylcytosine. Nat. Protoc. 2012, 7, 2159–2170.
Xue, L.; Yamazaki, H.; Ren, R.; Wanunu, M.; Ivanov, A. P.; Edel, J. B. Solid-state nanopore sensors. Nat. Rev. Mater. 2020, 5, 931–951.
Venkatesan, B. M.; Bashir, R. Nanopore sensors for nucleic acid analysis. Nat. Nanotechnol. 2011, 6, 615–624.
Dekker, C. Solid-state nanopores. Nat. Nanotechnol. 2007, 2, 209–215.
He, Y. H.; Tsutsui, M.; Zhou, Y.; Miao, X. S. Solid-state nanopore systems: From materials to applications. NPG Asia Mater. 2021, 13, 48.
Carlsen, A. T.; Zahid, O. K.; Ruzicka, J.; Taylor, E. W.; Hall, A. R. Interpreting the conductance blockades of DNA translocations through solid-state nanopores. ACS Nano 2014, 8, 4754–4760.
Li, J. L.; Gershow, M.; Stein, D.; Brandin, E.; Golovchenko, J. A. DNA molecules and configurations in a solid-state nanopore microscope. Nat. Mater. 2003, 2, 611–615.
Storm, A. J.; Chen, J. H.; Zandbergen, H. W.; Dekker, C. Translocation of double-strand DNA through a silicon oxide nanopore. Phys. Rev. E 2005, 71, 051903.
Storm, A. J.; Storm, C.; Chen, J. H.; Zandbergen, H.; Joanny, J. F.; Dekker, C. Fast DNA translocation through a solid-state nanopore. Nano Lett. 2005, 5, 1193–1197.
Wanunu, M.; Sutin, J.; McNally, B.; Chow, A.; Meller, A. DNA translocation governed by interactions with solid-state nanopores. Biophys. J. 2008, 95, 4716–4725.
Wanunu, M.; Dadosh, T.; Ray, V.; Jin, J. M.; McReynolds, L.; Drndić, M. Rapid electronic detection of probe-specific MicroRNAs using thin nanopore sensors. Nat. Nanotechnol. 2010, 5, 807–814.
Lin, Y.; Ying, Y. L.; Shi, X.; Liu, S. C.; Long, Y. T. Direct sensing of cancer biomarkers in clinical samples with a designed nanopore. Chem. Commun. 2017, 53, 11564–11567.
Rivas, F.; Zahid, O. K.; Reesink, H. L.; Peal, B. T.; Nixon, A. J.; DeAngelis, P. L.; Skardal, A.; Rahbar, E.; Hall, A. R. Label-free analysis of physiological hyaluronan size distribution with a solidstate nanopore sensor. Nat. Commun. 2018, 9, 1037.
He, L. Q.; Tessier, D. R.; Briggs, K.; Tsangaris, M.; Charron, M.; McConnell, E. M.; Lomovtsev, D.; Tabard-Cossa, V. Digital immunoassay for biomarker concentration quantification using solidstate nanopores. Nat. Commun. 2021, 12, 5348.
Wu, R. P.; Wang, Y. S.; Zhu, Z. T.; Yu, C. M.; Li, H.; Li, B. L.; Dong, S. J. Low-noise solid-state nanopore enhancing direct labelfree analysis for small dimensional assemblies induced by specific molecular binding. ACS Appl. Mater. Interfaces 2021, 13, 9482–9490.
Iqbal, S. M.; Akin, D.; Bashir, R. Solid-state nanopore channels with DNA selectivity. Nat. Nanotechnol. 2007, 2, 243–248.
Mussi, V.; Fanzio, P.; Repetto, L.; Firpo, G.; Stigliani, S.; Tonini, G. P.; Valbusa, U. “DNA-dressed nanopore” for complementary sequence detection. Biosens. Bioelectron. 2011, 29, 125–131.
Cai, S. L.; Pataillot-Meakin, T.; Shibakawa, A.; Ren, R.; Bevan, C. L.; Ladame, S.; Ivanov, A. P.; Edel, J. B. Single-molecule amplification-free multiplexed detection of circulating MicroRNA cancer biomarkers from serum. Nat. Commun. 2021, 12, 3515.
He, L. Q.; Karau, P.; Tabard-Cossa, V. Fast capture and multiplexed detection of short multi-arm DNA stars in solid-state nanopores. Nanoscale 2019, 11, 16342–16350.
Zhang, Y.; Gu, Z. D.; Zhao, J. B.; Shao, L. Y.; Kan, Y. J. Sequence-specific detection of DNA strands using a solid-state nanopore assisted by microbeads. Micromachines 2020, 11, 1097.
Carlsen, A. T.; Zahid, O. K.; Ruzicka, J. A.; Taylor, E. W.; Hall, A. R. Selective detection and quantification of modified DNA with solid-state nanopores. Nano Lett. 2014, 14, 5488–5492.
Arima, A.; Tsutsui, M.; Washio, T.; Baba, Y.; Kawai, T. Solid-state nanopore platform integrated with machine learning for digital diagnosis of virus infection. Anal. Chem. 2021, 93, 215–227.
Howarth, M.; Chinnapen, D. J. F.; Gerrow, K.; Dorrestein, P. C.; Grandy, M. R.; Kelleher, N. L.; El-Husseini, A.; Ting, A. Y. A monovalent streptavidin with a single femtomolar biotin binding site. Nat. Methods 2006, 3, 267–273.
Fairhead, M.; Krndija, D.; Lowe, E. D.; Howarth, M. Plug-and-play pairing via defined divalent streptavidins. J. Mol. Biol. 2014, 426, 199–214.
Zahid, O. K.; Wang, F.; Ruzicka, J. A.; Taylor, E. W.; Hall, A. R. Sequence-specific recognition of MicroRNAs and other short nucleic acids with solid-state nanopores. Nano Lett. 2016, 16, 2033–2039.
Zahid, O. K.; Zhao, B. S.; He, C.; Hall, A. R. Quantifying mammalian genomic DNA hydroxymethylcytosine content using solid-state nanopores. Sci. Rep. 2016, 6, 29565.
Wang, F.; Zahid, O. K.; Swain, B. E.; Parsonage, D.; Hollis, T.; Harvey, S.; Perrino, F. W.; Kohli, R. M.; Taylor, E. W.; Hall, A. R. Solid-state nanopore analysis of diverse DNA base modifications using a modular enzymatic labeling process. Nano Lett. 2017, 17, 7110–7116.
Zahid, O. K.; Rivas, F.; Wang, F.; Sethi, K.; Reiss, K.; Bearden, S.; Hall, A. R. Solid-state nanopore analysis of human genomic DNA shows unaltered global 5-hydroxymethylcytosine content associated with early-stage breast cancer. Nanomed.: Nanotechnol., Biol. Med. 2021, 35, 102407.
Sethi, K.; Dailey, G. P.; Zahid, O. K.; Taylor, E. W.; Ruzicka, J. A.; Hall, A. R. Direct detection of conserved viral sequences and other nucleic acid motifs with solid-state nanopores. ACS Nano 2021, 15, 8474–8483.
Kowalczyk, S. W.; Wells, D. B.; Aksimentiev, A.; Dekker, C. Slowing down DNA translocation through a nanopore in lithium chloride. Nano Lett. 2012, 12, 1038–1044.
Sha, J. J.; Shi, H. J.; Zhang, Y.; Chen, C.; Liu, L.; Chen, Y. F. Salt gradient improving signal-to-noise ratio in solid-state nanopore. ACS Sens. 2017, 2, 506–512.
Wanunu, M.; Morrison, W.; Rabin, Y.; Grosberg, A. Y.; Meller, A. Electrostatic focusing of unlabelled DNA into nanoscale pores using a salt gradient. Nat. Nanotechnol. 2010, 5, 160–165.
Grosberg, A. Y.; Rabin, Y. DNA capture into a nanopore: Interplay of diffusion and electrohydrodynamics. J. Chem. Phys. 2010, 133, 165102.
Kowalczyk, S. W.; Grosberg, A. Y.; Rabin, Y.; Dekker, C. Modeling the conductance and DNA blockade of solid-state nanopores. Nanotechnology 2011, 22, 315101.
Smeets, R. M. M.; Keyser, U. F.; Dekker, N. H.; Dekker, C. Noise in solid-state nanopores. Proc. Natl. Acad. Sci. USA 2008, 105, 417–421.
Hagerman, P. J. Flexibility of DNA. Annu. Rev. Biophys. Biophys. Chem. 1988, 17, 265–286.
Weber, J. A.; Baxter, D. H.; Zhang, S. L.; Huang, D. Y.; How Huang, K.; Jen Lee, M.; Galas, D. J.; Wang, K. The MicroRNA spectrum in 12 body fluids. Clin. Chem. 2010, 56, 1733–1741.
Portela, A.; Esteller, M. Epigenetic modifications and human disease. Nat. Biotechnol. 2010, 28, 1057–1068.
Yang, J. J.; Ferranti, D. C.; Stern, L. A.; Sanford, C. A.; Huang, J.; Ren, Z.; Qin, L. C.; Hall, A. R. Rapid and precise scanning helium ion microscope milling of solid-state nanopores for biomolecule detection. Nanotechnology 2011, 22, 285310.
Haynes, W. M.; Lide, D. R.; Bruno, T. J. CRC Handbook of Chemistry and Physics: A Ready-Reference Book of Chemical and Physical Data, 97th ed.; CRC Press: Boca Raton, FL, USA, 2016.
Acknowledgements
This project was supported by NIH awards (Nos. R21CA193067, R33CA246448, and P41EB020594). SS-nanopore fabrication was performed at the Rutgers University Laboratory for Surface Modification. We acknowledge the laboratory of Dr. Mark Howarth (Oxford University) for providing MS protein. We thank Mallory Smith for contributions to figures.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Rights and permissions
About this article
Cite this article
Wadsworth, I.D., Hall, A.R. Effects of symmetric and asymmetric salt conditions on a selective solid-state nanopore assay. Nano Res. 15, 9936–9942 (2022). https://doi.org/10.1007/s12274-022-4631-9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12274-022-4631-9