Abstract
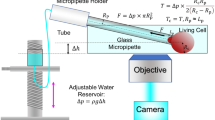
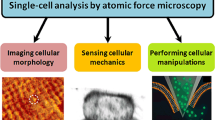
Fluidic force microscopy (FluidFM), which combines atomic force microscopy (AFM) with microchanneled cantilevers connected to a pressure controller, is a technique allowing the realization of force-sensitive nanopipette under aqueous conditions. FluidFM has unique advantages in simultaneous three-dimensional manipulations and mechanical measurements of biological specimens at the micro-/nanoscale. Over the past decade, FluidFM has shown its potential in biophysical assays particularly in the investigations at single-cell level, offering novel possibilities for discovering the underlying mechanisms guiding life activities. Here, we review the utilization of FluidFM to address biomechanical and biophysical issues in the life sciences. Firstly, the fundamentals of FluidFM are represented. Subsequently, the applications of FluidFM for biophysics at single-cell level are surveyed from several facets, including single-cell manipulations, single-cell force spectroscopy, and single-cell electrophysiology. Finally, the challenges and perspectives for future progressions are provided.

Article PDF
Similar content being viewed by others
Avoid common mistakes on your manuscript.
References
Altschuler, S. J.; Wu, L. F. Cellular heterogeneity: Do differences make a difference? Cell 2010, 141, 559–563.
Pennisi, E. Single-cell sequencing tackles basic and biomedical questions. Science 2012, 336, 976–977.
Pelkmans, L. Using cell-to-cell variability—A new era in molecular biology. Science 2012, 336, 425–426.
Ackermann, M. A functional perspective on phenotypic heterogeneity in microorganisms. Nat. Rev. Microbiol. 2015, 13, 497–508.
Raj, A.; van Oudenaarden, A. Nature, nurture, or chance: Stochastic gene expression and its consequences. Cell 2008, 135, 216–226.
Marusyk, A.; Almendro, V.; Polyak, K. Intra-tumour heterogeneity: A looking glass for cancer? Nat. Rev. Cancer 2012, 12, 323–334.
Burrell, R. A.; McGranahan, N.; Bartek, J.; Swanton, C. The causes and consequences of genetic heterogeneity in cancer evolution. Nature 2013, 501, 338–345.
Bedard, P. L.; Hansen, A. R.; Ratain, M. J.; Siu, L. L. Tumour heterogeneity in the clinic. Nature 2013, 501, 355–364.
Stuart, T.; Satija, R. Integrative single-cell analysis. Nat. Rev. Genet. 2019, 20, 257–272.
Wu, A. R.; Neff, N. F.; Kalisky, T.; Dalerba, P.; Treutlein, B.; Rothenberg, M. E.; Mburu, F. M.; Mantalas, G. L.; Sim, S.; Clarke, M. F. et al. Quantitative assessment of single-cell RNA-sequencing methods. Nat. Methods 2014, 11, 41–46.
Karemaker, I. D.; Vermeulen, M. Single-cell DNA methylation profiling: Technologies and biological applications. Trends Biotechnol. 2018, 36, 952–965.
Kang, C. C.; Yamauchi, K. A.; Vlassakis, J.; Sinkala, E.; Duncombe, T. A.; Herr, A. E. Single cell-resolution western blotting. Nat. Protoc. 2016, 11, 1508–1530.
Taheri-Araghi, S.; Brown, S. D.; Sauls, J. T.; McIntosh, D. B.; Jun, S. Sigle-cell physiology. Ann. Rev. Biophys. 2015, 44, 123–142
Patel, A. P.; Tirosh, I.; Trombetta, J. J.; Shalek, A. K.; Gillespie, S. M.; Wakimoto, H.; Cahill, D. P.; Nahed, B. V.; Curry, W. T.; Martuza, R. L. et al. Single-cell RNA-seq highlights intratumoral heterogeneity in primary glioblastoma. Science 2014, 344, 1396–1401.
Deng, Q. L.; Ramsköld, D.; Reinius, B.; Sandberg, R. Single-cell RNA-seq reveals dynamic, random monoallelic gene expression in mammalian cells. Science 2014, 343, 193–196.
Luquette, L. J.; Bohrson, C. L.; Sherman, M. A.; Park, P. J. Identification of somatic mutations in single cell DNA-seq using a spatial model of allelic imbalance. Nat. Commun. 2019, 10, 3908.
Dumont, S.; Prakash, M. Emergent mechanics of biological structures. Mol. Biol. Cell 2014, 25, 3461–3465.
Janmey, P. A.; McCulloch, C. A. Cell mechanics: Integrating cell responses to mechanical stimuli. Annu. Rev. Biomed. Eng. 2007, 9, 1–34.
Kim, D. H.; Wong, P. K.; Park, J.; Levchenko, A.; Sun, Y. Microengineered platforms for cell mechanobiology. Annu. Rev. Biomed. Eng. 2009, 11, 203–233.
Suresh, S. Biomechanics and biophysics of cancer cells. Acta Biomater. 2007, 3, 413–438.
Wirtz, D.; Konstantopoulos, K.; Searson, P. C. The physics of cancer: The role of physical interactions and mechanical forces in metastasis. Nat. Rev. Cancer 2011, 11, 512–522.
Mitchell, M. J.; Jain, R. K.; Langer, R. Engineering and physical sciences in oncology: Challenges and opportunities. Nat. Rev. Cancer 2017, 17, 659–675.
Yamauchi, K. A.; Herr, A. E. Subcellular western blotting of single cells. Microsyst. Nanoeng. 2017, 3, 16079.
Yu, W. B.; Sharma, S.; Gimzewski, J. K.; Rao, J. Y. Nanocytology as a potential biomarker for cancer. Biomark. Med. 2017, 11, 213–216.
Meister, A.; Gabi, M.; Behr, P.; Studer, P.; Vörös, J.; Niedermann, P.; Bitterli, J.; Polesel-Maris, J.; Liley, M.; Heinzelmann, H. et al. FluidFM: Combining atomic force microscopy and nanofluidics in a universal liquid delivery system for single cell applications and beyond. Nano Lett. 2009, 9, 2501–2507.
Binnig, G.; Quate, C. F.; Gerber, C. Atomic force microscope. Phys. Rev. Lett. 1986, 56, 930–933.
Li, M.; Xi, N.; Wang, Y. C.; Liu, L. Q. Advances in atomic force microscopy for single-cell analysis. Nano Res. 2019, 12, 703–718.
Dufrêne, Y. F.; Ando, T.; Garcia, R.; Alsteens, D.; Martinez-Martin, D.; Engel, A.; Gerber, C.; Muller, D. J. Imaging modes of atomic force microscopy for application in molecular and cell biology. Nat. Nanotechnol. 2017, 12, 295–307.
Krieg, M.; Fläschner, G.; Alsteens, D.; Gaub, B. M.; Roos, W. H.; Wuite, G. J. L.; Gaub, H. E.; Gerber, C.; Dufrêne, Y. F.; Muller, D. J. Atomic force microscopy-based mechanobiology. Nat. Rev. Phys. 2019, 1, 41–57.
Di Carlo, D. A mechanical biomarker of cell state in medicine. J. Lab. Autom. 2012, 17, 32–42.
Moeendarbary, E.; Harris, A. R. Cell mechanics: Principles, practices, and prospects. WIREs Syst. Biol. Med. 2014, 6, 371–388.
Li, M.; Dang, D.; Liu, L. Q.; Xi, N.; Wang Y. C. Atomic force microscopy in characterizing cell mechanics for biomedical applications: A review. IEEE Trans. NanoBiosci. 2017, 16, 523–540.
Harris, A. R.; Peter, L.; Bellis, J.; Baum, B.; Kabla, A. J.; Charras, G. T. Characterizing the mechanics of cultured cell monolayers. Proc. Natl. Acad. Sci. USA 2012, 109, 16449–16454.
Wu, P. H.; Aroush, D. R. B.; Asnacios, A.; Chen, W. C.; Dokukin, M. E.; Doss, B. L.; Durand-Smet, P.; Ekpenyong, A.; Guck, J.; Guz, N. V. et al. A comparison of methods to assess cell mechanical properties. Nat. Methods 2018, 15, 491–498.
Cross, S. E.; Jin, Y. S.; Rao, J. Y.; Gimzewski, J. K. Nanomechanical analysis of cells from cancer patients. Nat. Nanotechnol. 2007, 2, 780–783.
Plodinec, M.; Loparic, M.; Monnier, C. A.; Obermann, E. C.; Zanetti-Dallenbach, R.; Oertle, P.; Hyotyla, J. T.; Aebi, U.; Bentires-Alj, M.; Lim, R. Y. H.; Schoenenberger, C. A. The nanomechanical signature of breast cancer. Nat. Nanotechnol. 2012, 7, 757–765.
Gavara, N.; Chadwick, R. S. Determination of the elastic moduli of thin samples and adherent cells using conical atomic force microscope tips. Nat. Nanotechnol. 2012, 7, 733–736.
Garcia, R. Nanomechanical mapping of soft materials with the atomic force microscope: Methods, theory and applications. Chem. Soc. Rev. 2020, 49, 5850–5884.
Song, B.; Yang, R. G.; Xi, N.; Patterson, K. C.; Qu, C. G.; Lai, K. W. C. Cellular-level surgery using nano robots. J. Lab. Autom. 2012, 17, 425–434.
Han, S. W.; Nakamura, C.; Obataya, I.; Nakamura, N.; Miyake, J. A molecular delivery system by using AFM and nanoneedle. Biosens. Bioelectron. 2005, 20, 2120–2125.
Chen, X.; Kis, A.; Zettl, A.; Bertozzi, C. R. A cell nanoinjector based on carbon nanotubes. Proc. Natl. Acad. Sci. USA 2007, 104, 8218–8222.
Hochmuth, R. M. Micropipette aspiration of living cells. J. Biomech. 2000, 33, 15–22.
Zambelli, T.; Aebersold, M. J.; Behr, P.; Han, H.; Hirt, L.; Martinez, V.; Guillaume-Gentil, O.; Vörös, J. FluidFM: Development of the instrument as well as its applications for 2D and 3D lithography. In Open-Space Microfluidics: Concepts, Implementations, Applications. Delamarche, E.; Kaigala, G. V., Eds.; Wiley-VCH Verlag GmbH & Co. KGaA: Weinheim, 2018; pp 295–323.
Saha, P.; Duanis-Assaf, T.; Reches, M. Fundamentals and applications of FluidFM technology in single-cell studies. Adv. Mater. Interfaces 2020, 7, 2001115.
Li, M.; Xi, N.; Wang, Y. C.; Liu, L. Q. Atomic force microscopy for revealing micro/nanoscale mechanics in tumor metastasis: From single cells to microenvironmental cues. Acta Pharmacol. Sin. 2021, 42, 323–339.
Deladi, S.; Tas, N. R.; Berenschot, J. W.; Krijnen, G. J. M.; de Boer, M. J.; de Boer, J. H.; Peter, M.; Elwenspoek, M. C. Micromachined fountain pen for atomic force microscope-based nanopatterning. Appl. Phys. Lett. 2004, 85, 5361–5363.
Berenschot, E. J. W.; Burouni, N.; Schurink, B.; van Honschoten, J. W.; Sanders, R. G. P.; Truckenmuller, R.; Jansen, H. V.; Elwenspoek, M. C.; van Apeldoorn, A. A.; Tas, N. R. 3D nanofabrication of fluidic components by corner lithography. Small 2012, 5, 3823–3831.
Guillaume-Gentil, O.; Potthoff, E.; Ossola, D.; Franz, C. M.; Zambelli, T.; Vorholt, J. A. Force-controlled manipulation of single cells: From AFM to FluidFM. Trends Biotechnol. 2014, 32, 381–388.
Aramesh, M.; Forró, C.; Dorwling-Carter, L.; Lüchtefeld, I.; Schlotter, T.; Ihle, S. J.; Shorubalko, I.; Hosseini, V.; Momotenko, D.; Zambelli, T. et al. Localized detection of ions and biomolecules with a force-controlled scanning nanopore microscope. Nat. Nanotechnol. 2019, 14, 791–798.
Martinez, V.; Behr, P.; Drechsler, U.; Polesel-Maris, J.; Potthoff, E.; Vörös, J.; Zambelli, T. SU-8 hollow cantilevers for AFM cell adhesion studies. J. Micromech. Microeng. 2016, 26, 055006.
Helfricht, N.; Mark, A.; Dorwling-Carter, L.; Zambelli, T.; Papastavrou, G. Extending the limits of direct force measurements: Colloidal probes from sub-micron particles. Nanoscale 2017, 9, 9491–9501.
Guillaume-Gentil, O.; Mittelviefhaus, M.; Dorwling-Carter, L.; Zambelli, T.; Vorholt, J. A. FluidFM applications in single-cell biology. In Open-Space Microfluidics: Concepts, Implementations, Applications. Delamarche, E.; Kaigala, G. V., Eds.; Wiley-VCH Verlag GmbH & Co. KGaA: Weinheim, 2018; pp 325–354.
Konradi, R.; Acikgoz, C.; Textor, M. Polyoxazolines for nonfouling surface coatings-a direct comparison to the gold standard PEG. Macromol. Rapid Commun. 2012, 33, 1663–1676.
Weydert, S.; Zürcher, S.; Tanner, S.; Zhang, N.; Ritter, R.; Peter, T.; Aebersold, M. J.; Thompson-Steckel, G.; Forró, C. et al. Easy to apply polyoxazoline-based coating for precise and long-term control of neural patterns. Langmuir 2017, 33, 8594–8605.
Schlotter, T.; Weaver, S.; Forró, C.; Momotenko, D.; Vörös, J.; Zambelli, T.; Aramesh, M. Force-controlled formation of dynamic nanopores for single-biomolecule sensing and single-cell secretomics. ACS Nano 2020, 14, 12993–13003.
Guillaume-Gentil, O.; Zambelli, T.; Vorholt, J. A. Isolation of single mammalian cells from adherent cultures by fluidic force microscopy. Lab Chip 2014, 14, 402–414.
Martinez, V.; Forró, C.; Weydert, S.; Aebersold, M. J.; Dermutz, H.; Guillaume-Gentil, O.; Zambelli, T.; Vörös, J.; Demkó, L. Controlled single-cell deposition and patterning by highly flexible hollow cantilevers. Lab Chip 2016, 16, 1663–1674.
Collins, D. J.; Morahan, B.; Garcia-Bustos, J.; Doerig, C.; Plebanski, M.; Neild, A. Two-dimensional single-cell patterning with one cell per well driven by surface acoustic waves. Nat. Commun. 2015, 6, 8686.
Xue, X. F.; Sun, Y. B.; Resto-Irizarry, A. M.; Yuan, Y.; Yong, K. M. A.; Zheng, Y.; Weng, S. N.; Shao, Y.; Chai, Y. M.; Studer, L. et al. Mechanics-guided embryonic patterning of neuroectoderm tissue from human pluripotent stem cells. Nat. Mater. 2018, 17, 633–641.
Li, Q. S., Lee, G. Y. H., Ong, C. N., Lim, C. T. AFM indentation study of breast cancer cells. Biochem. Biophys. Res. Commun. 2008, 374, 609–613.
Li, M.; Liu, L. Q.; Xi, N.; Wang, Y. C.; Dong, Z. L.; Xiao, X. B.; Zhang, W. J. Atomic force microscopy imaging and mechanical properties measurement of red blood cells and aggressive cancer cells. Sci. China Life Sci. 2012, 55, 968–973.
Dimitriadis, E. K.; Horkay, F.; Maresca, J.; Kachar, B.; Chadwick, R. S. Determination of elastic moduli of thin layers of soft material using the atomic force microscope. Biophys. J. 2002, 82, 2798–2810.
Hu, H.; Shi, B.; Breslin, C. M.; Gignac, L.; Peng, Y. T. A sub-micron spherical atomic force microscopic tip for surface measurements. Langmuir 2020, 36, 7861–7867.
Dörig, P.; Ossola, D.; Truong, A. M.; Graf, M.; Stauffer, F.; Vörös, J.; Zambelli, T. Exchangeable colloidal AFM probes for the quantification of irreversible and long-term interactions. Biophys. J. 2013, 105, 463–472.
Lüchtefeld, I.; Bartolozzi, A.; Morales, J. M.; Dobre, O.; Basso, M.; Zambelli, T.; Vassalli, M. Elasticity spectra as a tool to investigate actin cortex mechanics. J. Nanobiotechnol. 2020, 18, 147.
Hinterdorfer, P.; Dufrêne, Y. F. Detection and localization of single molecular recognition events using atomic force microscopy. Nat. Methods 2006, 3, 347–355.
Stiefel, P.; Schmidt, F. I.; Dörig, P.; Behr, P.; Zambelli, T.; Vorholt, J. A.; Mercer, J. Cooperative vaccinia infection demonstrated at the single-cell level using FluidFM. Nano Lett. 2012, 12, 4219–4227.
Guillaume-Gentil, O.; Potthoff, E.; Ossola, D.; Dörig, P.; Zambelli, T.; Vorholt, J. A. Force-controlled fluidic injection into single cell nuclei. Small 2013, 9, 1904–1907.
Liu, H. J.; Wen, J.; Xiao, Y.; Liu, J.; Hopyan, S.; Radisic, M.; Simmons, C. A.; Sun, Y. In situ mechanical characterization of the cell nucleus by atomic force microscopy. ACS Nano 2014, 8, 3821–3828.
Guillaume-Gentil, O.; Grindberg, R. V.; Kooger, R.; Dorwling-Carter, L.; Martinez, V.; Ossola, D.; Pilhofer, M.; Zambelli, T.; Vorholt, J. A. Tunable single-cell extraction for molecular analyses. Cell 2016, 166, 506–516.
Guillaume-Gentil, O.; Rey, T.; Kiefer, P.; Ibáñez, A. J.; Steinhoff, R.; Brönnimann, R.; Dorwling-Carter, L.; Zambelli, T.; Zenobi, R.; Vorholt, J. A. Single-cell mass spectrometry of metabolites extracted from live cells by fluidic force microscopy. Anal. Chem. 2017, 89, 5017–5023.
Weaver, V. M. Cell and tissue mechanics: The new cell biology frontier. Mol. Biol. Cell 2017, 28, 1815–1818.
Gardel, M. L. Moving beyond molecular mechanisms. J. Cell Biol. 2015, 208, 143–145.
Helenius, J.; Heisenberg, C. P.; Gaub, H. E.; Muller, D. J. Single-cell force spectroscopy. J. Cell Sci. 2008, 121, 1785–1791.
Friedrichs, J.; Helenius, J.; Muller, D. J. Quantifying cellular adhesion to extracellular matrix components by single-cell force spectroscopy. Nat. Protoc. 2010, 5, 1353–1361.
Müller, D. J.; Dufrêne, Y. F. Atomic force microscopy: A nanoscopic window on the cell surface. Trends Cell Biol. 2011, 21, 461–469.
Dufrêne, Y. F. Atomic force microscopy and chemical force microscopy of microbial cells. Nat. Protoc. 2008, 3, 1132–1138.
Dehullu, J.; Vorholt, J. A.; Lipke, P. N.; Dufrene, Y. F. Fluidic force microscopy captures amyloid bonds between microbial cells. Trends Microbiol. 2019, 27, 728–730.
Potthoff, E.; Guillaume-Gentil, O.; Ossola, D.; Polesel-Maris, J.; LeibundGut-Landmann, S.; Zambelli, T.; Vorholt, J. A. Rapid and serial quantification of adhesion forces of yeast and mammalian cells. PLoS One 2012, 7, e52712.
Potthoff, E.; Franco, D.; D’Alessandro, V.; Starck, C.; Falk, V.; Zambelli, T.; Vorholt, J. A.; Poulikakos, D.; Ferrari, A. Toward a rational design of surface textures promoting endothelialization. Nano Lett. 2014, 14, 1069–1079.
McGrath, J. S.; Quist, J.; Seddon, J. R. T.; Lai, S. C. S.; Lemay, S. G.; Bridle, H. L. Deformability assessment of waterborne protozoa using a microfluidic-enabled force microscopy probe. PLoS One 2016, 11, e0150438.
Sankaran, S.; Jaatinen, L.; Brinkmann, J.; Zambelli, T.; Vörös, J.; Jonkheijm, P. Cell adhesion on dynamic supramolecular surfaces probed by fluid force microscopy-based single-cell force spectroscopy. ACS Nano 2017, 11, 3867–3874.
Jaatinen, L.; Young, E.; Hyttinen, J.; Vörös, J.; Zambelli, T.; Demkó, L. Quantifying the effect of electric current on cell adhesion studied by single-cell force spectroscopy. Biointerphases 2016, 11, 011004.
Sancho, A.; Vandersmissen, I.; Craps, S.; Luttun, A.; Groll, J. A new strategy to measure intercellular adhesion forces in mature cell-cell contacts. Sci. Rep. 2017, 7, 46152.
Cohen, N.; Sarkar, S.; Hondroulis, E.; Sabhachandani, P.; Konry, T. Quantification of intercellular adhesion forces measured by fluid force microscopy. Talanta 2017, 174, 409–413.
Ryma, M.; Blöhbaum, J.; Singh, R.; Sancho, A.; Matuszak, J.; Cicha, I.; Groll, J. Easy-to-prepare coating of standard cell culture dishes for cell-sheet engineering using aqueous solutions of poly (2-n-propyloxazoline). ACS Biomater. Sci. Eng. 2019, 5, 1509–1517.
Wysotzki, P.; Sancho, A.; Gimsa, J.; Groll, J. A comparative analysis of detachment forces and energies in initial and mature cell-material interaction. Colloids Surf B Biointerfaces 2020, 190, 110894.
Sztilkovics, M.; Gerecsei, T.; Peter, B.; Saftics, A.; Kurunczi, S.; Szekacs, I.; Szabo, B.; Horvath, R. Single-cell adhesion force kinetics of cell populations from combined label-free optical biosensor and robotic fluidic force microscopy. Sci. Rep. 2020, 10, 61.
Veerachamy, S.; Yarlagadda, T.; Manivasagam, G.; Yarlagadda, P. K. Bacterial adherence and biofilm formation on medical implants: A review. Proc. Inst. Mech. Eng. H J. Eng. Med. 2014, 228, 1083–1099.
Arciola, C. R.; Campoccia, D.; Montanaro, L. Implant infections: Adhesion, biofilm formation and immune evasion. Nat. Rev. Microbiol. 2018, 16, 397–409.
Dufrêne, Y. F. Sticky microbes: Forces in microbial cell adhesion. Trends Microbiol. 2015, 23, 376–382.
Potthoff, E.; Ossola, D.; Zambelli, T.; Vorholt, J. A. Bacterial adhesion force quantification by fluidic force microscopy. Nanoscale 2015, 7, 4070–4079.
Sprecher, K. S.; Hug, I.; Nesper, J.; Potthoff, E.; Mahi, M. A.; Sangermani, M.; Kaever, V.; Schwede, T.; Vorholt, J.; Jenal, U. Cohesive properties of the Caulobacter crescentus holdfast adhesin are regulated by a novel c-di-GMP effector protein. mBio 2017, 8, e00294–17.
Hoyer, L. L.; Cota, E. Candida albicans agglutinin-like sequence (Als) family vignettes: A review of Als protein structure and function. Front. Microbiol. 2016, 7, 280.
Dehullu, J.; Valotteau, C.; Herman-Bausier, P.; Garcia-Sherman, M.; Mittelviefhaus, M.; Vorholt, J. A.; Lipke, P. N.; Dufrene, Y. F. Fluidic force microscopy demonstrates that homophilic adhesion by Candida albicans Als proteins is mediated by amyloid bonds between cells. Nano Lett. 2019, 19, 3846–3853.
Hofherr, L.; Müller-Renno, C.; Ziegler, C. FluidFM as a tool to study adhesion forces of bacteria-optimization of parameters and comparison to conventional bacterial probe scanning force spectroscopy. PLoS One 2020, 15, e0227395.
Mittelviefhaus, M.; Müller, D. B.; Zambelli, T.; Vorholt, J. A. A modular atomic force microscopy approach reveals a large range of hydrophobic adhesion forces among bacterial members of the leaf microbiota. ISME J. 2019, 13, 1878–1882.
Mathelié-Guinlet, M.; Viela, F.; Viljoen, A.; Dehullu, J.; Dufrêne, Y. F. Single-molecule atomic force microscopy studies of microbial pathogens. Curr. Opin. Biomed. Eng. 2019, 12, 1–7.
Doll, K.; Yang, I.; Fadeeva, E.; Kommerein, N.; Szafrański, S. P.; der Wieden, G. B.; Greuling, A.; Winkel, A.; Chichkov, B. N.; Stumpp, N. S.; Stiesch, M. Liquid-infused structured titanium surfaces: Antiadhesive mechanism to repel Streptococcus oralis biofilms. ACS Appl. Mater. Interfaces 2019, 11, 23026–23038.
Dunlop, J.; Bowlby, M.; Peri, R.; Vasilyev, D.; Arias, R. High-throughput electrophysiology: An emerging paradigm for ion-channel screening and physiology. Nat. Rev. Drug Discov. 2008, 7, 358–368.
Becchetti, A.; Petroni, G.; Arcangeli, A. Ion channel conformations regulate integrin-dependent signaling. Trends Cell Biol. 2019, 29, 298–307.
Zaydman, M. A.; Silva, J. R.; Cui, J. M. Ion channel associated diseases: Overview of molecular mechanisms. Chem. Rev. 2012, 112, 6319–6333.
Chen, C. C.; Cang, C. L.; Fenske, S.; Butz, E.; Chao, Y. K.; Biel, M.; Ren, D. J.; Wahl-Schott, C.; Grimm, C. Patch-clamp technique to characterize ion channels in enlarged individual endolysosomes. Nat. Protoc. 2017, 12, 1639–1658.
Annecchino, L. A.; Morris, A. R.; Copeland, C. S.; Agabi, O. E.; Chadderton, P.; Schultz, S. R. Robotic automation of in vivo two-photon targeted whole-cell patch-clamp electrophysiology. Neuron 2017, 95, 1048–1055.e3.
Obergrussberger, A.; Goetze, T. A.; Brinkwirth, N.; Becker, N.; Friis, S.; Rapedius, M.; Haarmann, C.; Rinke-Weiβ, I.; Stölzle-Feix, S.; Brüggemann, A. et al. An update on the advancing high-throughput screening techniques for patch clamp-based ion channel screens: Implications for drug discovery. Expert Opin. Drug Discov. 2018, 13, 269–277.
Ossola, D.; Amarouch, M. Y.; Behr, P.; Vörös, J.; Abriel, H.; Zambelli, T. Force-controlled patch clamp of beating cardiac cells. Nano Lett. 2015, 15, 1743–1750.
Saotome, K.; Murthy, S. E.; Kefauver, J. M.; Whitwam, T.; Patapoutian, A.; Ward, A. B. Structure of the mechanically activated ion channel Piezo1. Nature 2018, 554, 481–486.
Nonomura, K.; Lukacs, V.; Sweet, D. T.; Goddard, L. M.; Kanie, A.; Whitwam, T.; Ranade, S. S.; Fujimori, T.; Kahn, M. L.; Patapoutian, A. Mechanically activated ion channel PIEZO1 is required for lymphatic valve formation. Proc. Natl. Acad. Sci. USA 2018, 115, 12817–12822.
Cox, C. D.; Bavi, N.; Martinac, B. Biophysical principles of ion-channel-mediated mechanosensory transduction. Cell Rep. 2019, 29, 1–12.
Li, M.; Dang, D.; Xi, N.; Wang, Y. C.; Liu, L. Q. Nanoscale imaging and force probing of biomolecular systems using atomic force microscopy: From single molecules to living cells. Nanoscale 2017, 9, 17643–17666.
Müller, D. J.; Dufrêne, Y. F. Force nanoscopy of living cells. Curr. Biol. 2011, 21, R212–R216.
Seifert, J.; Rheinlaender, J.; Novak, P.; Korchev, Y. E.; Schäffer, T. E. Comparison of atomic force microscopy and scanning ion conductance microscopy for live cell imaging. Langmuir 2015, 31, 6807–6813.
Ando, T. High-speed atomic force microscopy and its future prospects. Biophys. Rev. 2018, 10, 285–292.
Ossola, D.; Dorwling-Carter, L.; Dermutz, H.; Behr, P.; Vörös, J.; Zambelli, T. Simultaneous scanning ion conductance microscopy and atomic force microscopy with microchanneled cantilevers. Phys. Rev. Lett. 2015, 115, 238103.
Yasui, M.; Hiroshima, M.; Kozuka, J.; Sako, Y.; Ueda, M. Automated single-molecule imaging in living cells. Nat. Commun. 2018, 9, 3061.
Sako, Y.; Minoghchi, S.; Yanagida, T. Single-molecule imaging of EGFR signalling on the surface of living cells. Nat. Cell Biol. 2000, 2, 168–172.
Elf, J.; Li, G. W.; Xie, X. S. Probing transcription factor dynamics at the single-molecule level in a living cell. Science 2007, 316, 1191–1194.
Tokunaga, M.; Imamoto, N.; Sakata-Sogawa, K. Highly inclined thin illumination enables clear single-molecule imaging in cells. Nat. Methods 2008, 5, 159–161.
Kodera, N.; Yamamoto, D.; Ishikawa, R.; Ando, T. Video imaging of walking myosin V by high-speed atomic force microscopy. Nature 2010, 468, 72–76.
Ying, Y. L.; Hu, Y. X.; Gao, R.; Yu, R. J.; Gu, Z.; Lee, L. P.; Long, Y. T. Asymmetric nanopore electrode-based amplification for electron transfer imaging in live cells. J. Am. Chem. Soc. 2018, 140, 5385–5392.
Li, M.; Xi, N.; Wang, Y. C.; Liu, L. Q. In situ high-resolution AFM imaging and force probing of cell culture medium-forming nanogranular surfaces for cell growth. IEEE Trans. NanoBiosci. 2020, 19, 385–393.
Li, L.; Guo, W.; Yan, Y. Z.; Lee, S.; Wang, T. Label-free superresolution imaging of adenoviruses by submerged microsphere optical nanoscopy. Light Sci. Appl. 2013, 2, e104.
Lekka, M.; Pogoda, K.; Gostek, J.; Klymenko, O.; Prauzner-Bechcicki, S.; Wiltowska-Zuber, J.; Jaczewska, J.; Lekki, J.; Stachura, Z. Cancer cell recognition — mechanical phenotype. Micron 2012, 43, 1259–1266.
Li, M.; Xi, N.; Wang, Y. C.; Liu, L. Q. Atomic force microscopy in probing tumor physics for nanomedicine. IEEE Trans. Nanotechnol. 2019, 15, 83–113.
Tian, Y. M.; Li, J. H.; Cai, M. J.; Zhao, W. D.; Xu, H. J.; Liu, Y.; Wang, H. D. High resolution imaging of mitochondrial membranes by in situ atomic force microscopy. RSC Adv. 2013, 3, 708–712.
Wang, X.; Liu, H. J.; Zhu, M.; Cao, C. H.; Xu, Z. S.; Tsatskis, Y.; Lau, K.; Kuok, C.; Filleter, T.; McNeill, H. et al. Mechanical stability of the cell nucleus-roles played by the cytoskeleton in nuclear deformation and strain recovery. J. Cell Sci. 2018, 131, jcs209627.
Stewart, J. B.; Chinnery, P. F. Extreme heterogeneity of human mitochondrial DNA from organelles to populations. Nat. Rev. Genet. 2021, 22, 106–118.
Dan, K.; Veetil, A. T.; Chakraborty, K.; Krishnan, Y. DNA nanodevices map enzymatic activity in organelles. Nat. Nanotechnol. 2019, 14, 252–259.
Acknowledgements
This work was supported by the China Scholarship Council (CSC) (No. 202004910157). M. Li is thankful for the support from the National Natural Science Foundation of China (Nos. 61922081 and 61873258), the Key Research Program of Frontier Sciences CAS (No. ZDBS-LY-JSC043), the Youth Innovation Promotion Association CAS (No. 2017243), and the LiaoNing Revitalization Talents Program (No. XLYC1907072).
Funding
Open Access funding provided by Swiss Federal Institute of Technology Zurich
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made.
The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder.
To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Li, M., Liu, L. & Zambelli, T. FluidFM for single-cell biophysics. Nano Res. 15, 773–786 (2022). https://doi.org/10.1007/s12274-021-3573-y
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12274-021-3573-y