Abstract
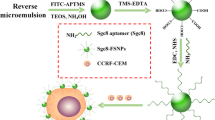
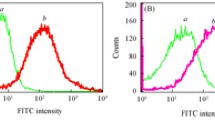
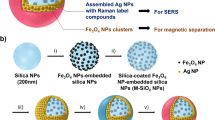
Early and accurate diagnosis and treatment of cancer depend on rapid, sensitive, and selective detection of tumor cells. Current diagnosis of cancers, especially leukemia, relies on histology and flow cytometry using single dye-labeled antibodies. However, this combination may not lead to high signal output, which can hinder detection, especially when the probes have relatively weak affinities or when the receptor is expressed in a low concentration on the target cell surface. To solve these problems, we have developed a novel method for sensitive and rapid detection of cancer cells using dye-doped silica nanoparticles (NPs) which increases detection sensitivity in flow cytometry analyses between 10- and 100-fold compared to standard methods. Our NPs are ∼60 nm in size and can encapsulate thousands of individual dye molecules within their matrix. We have extensively investigated surface modification strategies in order to make the NPs suitable for selective detection of cancer cells using flow cytometry. The NPs are functionalized with polyethylene glycol (PEG) to prevent nonspecific interactions and with neutravidin to allow universal binding with biotinylated molecules. By virtue of their reliable and selective detection of target cancer cells, these NPs have demonstrated their promising usefulness in conventional flow cytometry. Moreover, they have shown low background signal, high signal enhancement, and efficient functionalization, either with antibody- or aptamer-targeting moieties.

Article PDF
Similar content being viewed by others
Avoid common mistakes on your manuscript.
References
Prince, H. E.; Arens, L.; Kleinman, S. H. CD4 and CD8 subsets defined by dual-color cytofluorometry which distinguish symptomatic from asymptomatic blood donors seropositive for human immunodeficiency virus. Diagn. Clin. Immunol. 1987, 5, 188–193.
Craig, F. E. Flow cytometric evaluation of B-cell lymphoid neoplasms. Clin. Lab. Med. 2007, 27, 487–512.
Donnenberg, A. D.; Donnenberg, V. S. Rare-event analysis in flow cytometry. Clin. Lab. Med. 2007, 27, 627–652.
Niemeyer, C. M. Nanoparticles, proteins, and nucleic acids: Biotechnology meets materials science. Angew. Chem. Int. Ed. 2001, 40, 4128–4158.
Wang, L.; Wang, K.; Santra, S.; Zhao, X.; Hilliard, L. R.; Smith, J. E.; Wu, Y.; Tan, W. Watching silica nanoparticles glow in the biological world. Anal. Chem. 2006, 78, 646–654.
Holm, B. A.; Bergey, E. J.; De T; Rodman, D. J.; Kapoor, R.; Levy, L.; Friend, C. S.; Prasad, P. N. Nanotechnology in biomedical applications. Mol. Cryst. Liq. Cryst. 2002, 374, 589–598.
He, X.; Duan, J.; Wang, K.; Tan, W.; Lin, X.; He, C. A novel fluorescent label based on organic dye-doped silica nanoparticles for HepG liver cancer cell recognition. J. Nanosci. Nanotechnol. 2004, 4, 585–589.
Herr, J. K.; Smith, J. E.; Medley, C. D.; Shangguan, D.; Tan, W. Aptamer-conjugated nanoparticles for selective collection and detection of cancer cells. Anal. Chem. 2006, 78, 2918–2924.
Zhao, X.; Hilliard, L. R.; Mechery, S. J.; Wang, Y.; Bagwe, R. P.; Jin, S.; Tan, W. A rapid bioassay for single bacterial cell quantitation using bioconjugated nanoparticles. Proc. Natl. Acad. Sci. USA 2004, 101, 15027–15032.
Shangguan, D.; Li, Y.; Tang, Z.; Cao, Z. C.; Chen, H. W.; Mallikaratchy, P.; Sefah, K.; Yang, C. J.; Tan, W. Aptamers evolved from live cells as effective molecular probes for cancer study. Proc. Natl. Acad. Sci. 2006, 103, 11838–11843.
Tang, Z.; Shangguan, D.; Wang, K.; Shi, H.; Sefah, K.; Mallikaratchy, P.; Chen, H. W.; Li, Y.; Tan, W. Selection of aptamers for molecular recognition and characterization of cancer cells. Anal. Chem. 2007, 79, 4900–4907.
Chattopadhyay, P. K.; Price, D. A.; Harper, T. F.; Betts, M. R.; Yu, J.; Gostick, E.; Perfetto, S. P.; Goepfert, P.; Koup, R. A.; De Rosa, S. C.; Bruchez, M. P.; Roederer, M. Quantum dot semiconductor nanocrystals for immunophenotyping by polychromatic flow cytometry. Nat. Med. 2006, 12, 972–977.
Stöber, W.; Fink, A.; Bohn, E. Controlled growth of monodisperse silica spheres in the micron size range. J. Colloid Interf. Sci. 1968, 26, 62–69.
Bagwe, R. P.; Zhao, X.; Tan, W. Bioconjugated luminescent nanoparticles for biological applications. J. Disper. Sci. Technol. 2003, 24, 453–464.
Bagwe, R. P.; Hilliard, L. R.; Tan, W. Surface modification of silica nanoparticles to reduce aggregation and nonspecific binding. Langmuir 2006, 22, 4357–4362.
Hilliard, L. R.; Zhao, X.; Tan, W. Immobilization of oligonucleotides onto silica nanoparticles for DNA hybridization studies. Anal. Chim. Acta 2002, 470, 51–56.
Xu, H.; Yan, F.; Monson, E. E.; Kopelman, R. Room-temperature preparation and characterization of poly(ethylene glycol)-coated silica nanoparticles for biomedical applications. J. Biomed. Mater. Res. 2003, 66A, 870–879.
Gao, X.; Cui, Y.; Levenson, R. M.; Chung, L. W. K.; Shuming, N. In vivo cancer targeting and imaging with semiconductor quantum dots. Nat. Biotechnol. 2004, 22, 969–976.
Liu, Y.; Shipton, M. K.; Ryan, J.; Kaufman, E. D.; Franzen, S.; Feldheim, D. L. Synthesis, stability, and cellular internalization of gold nanoparticles containing mixed peptide-poly(ethylene glycol) monolayers. Anal. Chem. 2007, 79, 2221–2229.
van Blaaderen, A.; Vrij, A. Synthesis and characterization of colloidal dispersions of fluorescent, monodisperse silica spheres. Langmuir 1992, 8, 2921–2931.
Wang, L.; Tan, W. Multicolor FRET silica nanoparticles by single wavelength excitation. Nano Lett. 2006, 6, 84–88.
Santra, S.; Wang, K.; Tapec, R.; Tan, W. Development of novel dye-doped silica nanoparticles for biomarker application. J. Biomed. Opt. 2001, 6, 160–166.
Brussaard, C. P. D.; Marie, D.; Bratbak, G. Flow cytometric detection of viruses. J. Virol. Methods 2000, 85, 175–182.
Shangguan, D.; Cao, Z. C.; Li, Y.; Tan, W. Aptamers evolved from cultured cancer cells reveal molecular differences of cancer cells in patient samples. Clin. Chem. 2007, 53, 1153–1155.
Shangguan, D.; Cao, Z.; Meng, L.; Mallikaratchy, P.; Sefah, K.; Wang, H.; Li, Y.; Tan, W. Cell-specific aptamer probes for membrane protein elucidation in cancer cells. J. Proteome Res. 2008, 7, 2133–2139.
Xiao, Z.; Shangguan, D.; Cao, Z.; Fang, X.; Tan, W. Cellspecific internalization study of an aptamer from whole cell selection. Chem. Eur. J. 2008, 14, 1769–1775.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Rights and permissions
Open Access This is an open access article distributed under the terms of the Creative Commons Attribution Noncommercial License ( https://creativecommons.org/licenses/by-nc/2.0 ), which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
About this article
Cite this article
Estévez, M.C., O’Donoghue, M.B., Chen, X. et al. Highly fluorescent dye-doped silica nanoparticles increase flow cytometry sensitivity for cancer cell monitoring. Nano Res. 2, 448–461 (2009). https://doi.org/10.1007/s12274-009-9041-8
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12274-009-9041-8