Abstract
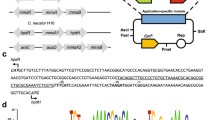
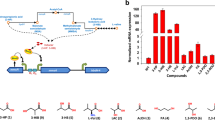
Here, we characterized a gene expression system induced by 3-hydroxypropionic acid (3-HP) in Pseudomonas denitrificans. The system consists of a putative LysR-type transcriptional regulator (LTTR) encoded by hpdR and a LTTR-responsive promoter that positively controls the expression of hpdH, encoding 3-HP dehydrogenase in the presence of 3-HP. In the hpdH-responsive promoter region, two operators exhibiting dyad symmetry and designated O1 and O2 and centered at the −73 and −30 positions, respectively, were located upstream of the hpdH transcription start site. When either O1, O2, or both regions were mutated, the inducibility by the HpdR-3-HP complex was significantly reduced or completely removed, indicating that both sites are required for transcriptional activation. The HpdR protein and its operator sites on hpdH were highly specific for each other, and did not engage in cross-talk with another similar 3-HP-inducible system that is also present in P. denitrificans. This 3-HP-inducible promoter system should be useful in developing biosensors for 3-HP, and/or dynamic gene expression systems for metabolic engineering purposes.
Similar content being viewed by others
References
Ongley, S. E., X. Bian, B. A. Neilan, and R. Müller (2013) Recent advances in the heterologous expression of microbial natural product biosynthetic pathways. Nat. Prod. Rep. 30: 1121–1138.
Kumar, V., S. Ashok, and S. Park (2013) Recent advances in biological production of 3-hydroxypropionic acid. Biotechnol. Adv. 31: 945–961.
Ashok, S., S. M. Raj, Y. Ko, M. Sankaranarayanan, S. Zhou, V. Kumar, and S. Park (2013) Effect of puuC overexpression and nitrate addition on glycerol metabolism and anaerobic 3-hydroxypropionic acid production in recombinant Klebsiella pneumoniae ΔglpKΔdhaT. Metab. Eng. 15: 10–24.
Ashok, S., M. Sankaranarayanan, Y. Ko, K. E. Jae, S. K. Ainala, V. Kumar, and S. Park (2013) Production of 3-hydroxypropionic acid from glycerol by recombinant Klebsiella pneumoniae ΔdhaTΔyqhD which can produce vitamin B12 naturally. Biotechnol. Bioeng. 110: 511–524.
Zhou, S., S. Ashok, Y. Ko, D. M. Kim, and S. Park (2014) Development of a deletion mutant of Pseudomonas denitrificans that does not degrade 3-hydroxypropionic acid. Appl. Microbiol. Biotechnol. 98: 4389–4398.
Zhou, S., C. Catherine, C. Rathnasingh, A. Somasundar, and S. Park (2013) Production of 3-hydroxypropionic acid from glycerol by recombinant Pseudomonas denitrificans. Biotechnol. Bioeng. 110: 3177–3187.
Lim, H. G., M. H. Noh, J. H. Jeong, S. Park, and G. Y. Jung (2016) Optimum rebalancing of the 3-hydroxypropionic acid production pathway from glycerol in Escherichia coli. ACS Synth. Biol. 5: 1247–1255.
Kwak, S., Y. C. Park, and J. H. Seo (2013) Biosynthesis of 3-hydroxypropionic acid from glycerol in recombinant Escherichia coli expressing Lactobacillus brevis dhaB and dhaR gene clusters and E. coli K-12 aldH. Bioresour. Technol. 135: 432–439.
Kim, K., S. K. Kim, Y. C. Park, and J. H. Seo (2014) Enhanced production of 3-hydroxypropionic acid from glycerol by modulation of glycerol metabolism in recombinant Escherichia coli. Bioresour. Technol. 156: 170–175.
Ko, Y., S. Ashok, E. Seol, S. K. Ainala, and S. Park (2015) Deletion of putative oxidoreductases from Klebsiella pneumoniae J2B could reduce 1,3-propanediol during the production of 3-hydroxypropionic acid from glycerol. Biotechnol. Bioprocess Eng. 20: 834–843.
Kumar, V., M. Sankaranarayanan, M. Durgapal, S. Zhou, Y. Ko, S. Ashok, R. Sarkar, and S. Park (2013) Simultaneous production of 3-hydroxypropionic acid and 1,3-propanediol from glycerol using resting cells of the lactate dehydrogenase-deficient recombinant Klebsiella pneumoniae overexpressing an aldehyde dehydrogenase. Bioresour. Technol. 135: 555–563.
Raj, S. M., C. Rathnasingh, J. E. Jo, and S. Park (2008) Production of 3-hydroxypropionic acid from glycerol by a novel recombinant Escherichia coli BL21 strain. Process Biochem. 43: 1440–1446.
Raj, S. M, C. Rathnasingh, W. C. Jung, E. Selvakumar, and S. Park (2010) A Novel NAD+-dependent aldehyde dehydrogenase encoded by the puuC gene of Klebsiella pneumoniae DSM 2026 that utilizes 3-hydroxypropionaldehyde as a substrate. Biotechnol. Bioprocess Eng. 15: 131–138.
Rathnasingh, C., S. M. Raj, Y. Lee, C. Catherine, S. Ashok, and S. Park (2012) Production of 3-hydroxypropionic acid via malonyl-CoA pathway using recombinant Escherichia coli strains. J. Biotechnol. 157: 633–640.
Sankaranarayanan, M., S. Ashok, and S. Park (2014) Production of 3-hydroxypropionic acid from glycerol by acid tolerant Escherichia coli. J. Ind. Microbiol. Biotechnol. 41: 1039–1050.
Zhou, S., S. K. Ainala, E. Seol, T. T. Nguyen, and S. Park (2015) Inducible gene expression system by 3-hydroxypropionic acid. Biotechnol. Biofuels. 8: 169.
Zhou, S., S. M. Raj, S. Ashok, S. Edwardraja, S. G. Lee, and S. Park (2013) Cloning, expression and characterization of 3-hydroxyisobutyrate dehydrogenase from Pseudomonas denitrificans ATCC 13867. PLoS One. 8: e62666.
Hoffmann, J. and J. Altenbuchner (2015) Functional characterization of the mannitol promoter of Pseudomonas fluorescens DSM 50106 and its application for a mannitol-inducible expression system for Pseudomonas putida KT2440. PLoS One. 10: e0133248.
Maddocks, S. E. and P. C. F. Oyston (2008) Structure and function of the LysR-type transcriptional regulator (LTTR) family proteins. Microbiology (Reading). 154: 3609–3623.
Schell, M. A. (1993) Molecular biology of the LysR family of transcriptional regulators. Annu. Rev. Microbiol. 47: 597–626.
Rhee, K. Y., M. Opel, E. Ito, S. Hung, S. M. Arfin, and G. W. Hatfield (1999) Transcriptional coupling between the divergent promoters of a prototypic LysR-type regulatory system, the ilvYC operon of Escherichia coli. Proc. Natl. Acad. Sci. USA. 96: 14294–14299.
Lochowska, A., R. Iwanicka-Nowicka, D. Plochocka, and M. M. Hryniewicz (2001) Functional dissection of the LysR-type CysB transcriptional regulator. Regions important for DNA binding, inducer response, oligomerization, and positive control. J. Biol. Chem. 276: 2098–2107.
Hanko, E. K. R., N. P. Minton, and N. Malys (2017) Characterisation of a 3-hydroxypropionic acid-inducible system from Pseudomonas putida for orthogonal gene expression control in Escherichia coli and Cupriavidus necator. Sci. Rep. 7: 1724.
Rhee, K. Y., D. F. Senear, and G. W. Hatfield (1998) Activation of gene expression by a ligand-induced conformational change of a protein-DNA complex. J. Biol. Chem. 273: 11257–11266.
Dillon, S. C., E. Espinosa, K. Hokamp, D. W. Ussery, J. Casadesus, and C. J. Dorman (2012) LeuO is a global regulator of gene expression in Salmonella enterica serovar Typhimurium. Mol. Microbiol. 85: 1072–1089.
Samarasinghe, S., M. S. El-Robh, D. C. Grainger, W. Zhang, P. Soultanas, and S. J. W. Busby (2008) Autoregulation of the Escherichia coli melR promoter: repression involves four molecules of MelR. Nucleic Acids Res. 36: 2667–2676.
Nguyen, N. H., J. R. Kim, and S. Park (2019) Development of biosensor for 3-hydroxypropionic acid. Biotechnol. Bioprocess Eng. 24: 109–118.
Rogers, J. K. and G. M. Church (2016) Genetically encoded sensors enable real-time observation of metabolite production. Proc. Natl. Acad. Sci. USA. 113: 2388–2393.
Rogers, J. K., N. D. Taylor, and G. M. Church (2016) Biosensor-based engineering of biosynthetic pathways. Curr. Opin. Biotechnol. 42: 84–91.
Ashok, S., S. M. Raj, C. Rathnasingh, and S. Park (2011) Development of recombinant Klebsiella pneumoniae ΔdhaT strain for the co-production of 3-hydroxypropionic acid and 1,3-propanediol from glycerol. Appl. Microbiol. Biotechnol. 90: 1253–1265.
Winsor, G. L., E. J. Griffiths, R. Lo, B. K. Dhillon, J. A. Shay, and F. S. L. Brinkman (2016) Enhanced annotations and features for comparing thousands of Pseudomonas genomes in the Pseudomonas genome database. Nucleic Acids Res. 44: D646–D653.
Nguyen, N. H., S. K. Ainala, S. Zhou, and S. Park (2019) A novel 3-hydroxypropionic acid-inducible promoter regulated by the LysR-type transcriptional activator protein MmsR of Pseudomonas denitrificans. Sci. Rep. 9: 5333.
Studer, G., C. Rempfer, A. M. Waterhouse, R. Gumienny, J. Haas, and T. Schwede (2020) QMEANDisCo — distance constraints applied on model quality estimation. Bioinformatics. 36: 1765–1771.
Acknowledgments
This study was financially supported by a grant from the C1 Gas Refinery Program (NRF-2017M3D3A1A01036927-2) through the National Research Foundation of Korea (NRF) funded by the Ministry of Science and ICT. Trinh Thi Nguyen was financially supported by the BK21 Plus Program of Pusan National University.
The authors declare no conflict of interest.
Neither ethical approval nor informed consent was required for this study.
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher’s Note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Co-first authors
Electronic Supplementary Material
Rights and permissions
About this article
Cite this article
Nguyen, T.T., Nguyen, N.H., Kim, Y. et al. In vivo Characterization of the Inducible Promoter System of 3-hydroxypropionic Dehydrogenase in Pseudomonas denitrificans. Biotechnol Bioproc E 26, 612–620 (2021). https://doi.org/10.1007/s12257-020-0291-3
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12257-020-0291-3