Abstract
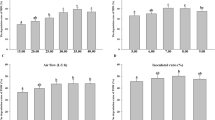
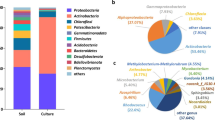
A native microbial consortium capable of degrading hydrocarbons was employed as an inoculum source in a sequencing batch reactor (SBR) using molasses as a carbon source. The microbial biomass in the SBR was able to grow in the presence of molasses, degrading 88% of the reducing sugar. Moreover, the consortium produced in the SBR was capable of maintaining 75% of the capacity for biodegradation of oil with respect to the original capacity of the native microbial consortium. Monitoring of the microbial population structure was accomplished using PCR-DGGE. The results indicated that the microbial populations grown in molasses were stable during crude oil degradation, as judged by comparison to the population structure of the native microbial consortium. The results obtained demonstrated that molasses could be used as a carbon source to promote the growth of biomass with oildegrading capacity.
Similar content being viewed by others
References
Wang, X., X. Wang, M. Liu, Y. Bu, J. Zhang, J. Chen, and J. Zhao (2015) Adsorption synergic biodegradation of diesel oil in synthetic seawater by acclimated strains immobilized on multifunctional materials. Mar. Pollut Bull. 92: 195–200.
Mariano, A. J., V. H. Kourafalou, A. Srinivasan, H. Kang, G. R. Halliwell, E. H. Ryan, and M. Roffer (2011) On the modeling of the 2010 gulf of México oil spill. Dynam. Atmos. Oceans 52: 322–340.
Ghazali, F. M., R. N. Z. A. Rahman, A. B. Salleh, and M. Basri (2004) Biodegradation of hydrocarbons in soil by microbial consortium. Int. Biodeterior. Biodegrad. 54: 61–67.
Sathishkumar, M., A. R. Binupriya, S. H. Baik, and S. E. Yun (2008) Biodegradation of crude oil by individual bacterial strains and a mixed bacterial consortium isolated from hydrocarbon contaminated areas. CLEAN-Soil, Air, Water 36: 92–96.
Gojgic-Cvijovic, G. D., J. S. Milic, T. M. Solevic, V. P. Beskoski, M. V. Ilic, L. S. Djokic, T. M. Narancic, and M. M. Vrvic (2012) Biodegradation of petroleum sludge and petroleum polluted soil by a bacterial consortium: a laboratory study. Biodegrad. 23: 1–14.
Ambujom, S. (2001) Studies on composition and stability of a large membered bacterial consortium degrading phenol. Microbiol. Res. 156: 293–302.
Kurachi, K., R. Hosokawa, M. Takahashi, and H. Okuyama (2014) The potential of glycerol in freezing preservation of turbine oil-degrading bacterial consortium and the ability of the revised consortium to degrade petroleum wastes. Int. Biodeterior. Biodegrad. 88: 77–82.
Wang, Z. Y., Y. Xu, H. -Y. Wang, J. Zhao, D. M. Gao, F. M. Li, and B. Xing (2012) Biodegradation of crude oil in contaminated soils by free and immobilized microorganisms. Pedosphere 22: 717–725.
Lin, M., Y. Liu, W. Chen, H. Wang, and X. Hu (2014) Use of bacteria-immobilized cotton fibers to absorb and degrade crudeoil. Int. Biodeterior. Biodegrad. 88: 8–12.
Mohan, S. V., G. Mohanakrishna, S. S. Reddy, B. D. Raju, K. S. R. Rao, and P. N. Sarma (2008) Self-immobilization of acidogenic mixed consortia on mesoporous material (SBA-15) and activated carbon to enhance fermentative hydrogen production. Int. J. Hydrogen Energy 33: 6133–6142.
Teclu, D., G. Tivchev, M. Laing, and M. Wallis (2009) Determination of the elemental composition of molasses and its suitability as carbon source for growth of sulphate-reducing bacteria. J. Hazard. Mater. 161: 1157–1165.
Gomes, E. B., R. F. Silva, A. S. Rosado, and J. Pereira (2010) Biotreatment of diesel waste by sequencing batch bioreactor operation mode (SBR). Inter. Biodeterior. Biodegrad. 64: 413–417.
Jalali, S., J. Shayegan, and S. Rezasoltani (2015) Rapid start-up and improvement of granulation in SBR. J. Environ. Health. Sci. Eng. 13: 1–11.
Venkata Mohan, S., S. V. Raghavulu, R. K. Goud, S. Srikanth, V. L. Babu, and P. N. Sarma (2010) Microbial diversity analysis of long term operated biofilm configured anaerobic reactor producing biohydrogen from wastewater under diverse conditions. Int. J. Hydrogen Energy 35: 12208–12215.
Muyzer, G., E. C. de Waal, and A. G. Uitterlinden (1993) Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl. Environ. Microbiol. 59: 695–700.
Hesham, A. E. -L., R. Qi, and M. Yang (2011) Comparison of bacterial community structures in two systems of a sewage treatment plant using PCR-DGGE analysis. J. Environ Sci. 23: 2049–2054.
Canul-Chan, M., N. Estrella-Gómez, A. Zepeda, D. Cabañas-Vargas, and R. Rojas-Herrera (2014) A protocol for Metagenomic RNA extraction from bacterial consortium in the presence of crude oil. Rom. Biotech. Lett. 19: 8910–8915.
Bushnell, L. D. and H. F. Haas (1941) The utilization of certain hydrocarbons by microorganisms. J. Bacteriol. 41: 653–673.
Rojas-Herrera, R., J. Narváez-Zapata, M. Zamudio-Maya, and M. Mena-Martínez (2008) A simple silica-based method for metagenomic DNA extraction from soil and sediments. Mol. Biotech. 40: 13–17.
Miller, G. L. (1959) Use of dinitrosalicyclic acid regeant for determination of reducing sugar. Anal. Chem. 31: 426–428.
Peterson, G. I. (1977) A simplification of the protein assay method of Lowry et al. which is more generally applicable. Anal. Biochem. 83: 346–356.
Deppe, U., H. H. Richnow, W. Michaelis, and G. Antranikian (2005) Degradation of crude oil by an arctic microbial consortium. Extremophiles 9: 461–470.
Antizar-Ladislao, B., K. Spanova, A. J. Beck, and N. J. Russell (2008) Microbial community structure changes during bioremediation of PAHs in an aged coal-tar contaminated soil by in-vessel composting. Int. Biodeterior. Biodegrad. 61: 357–364.
Kermanshahipour, A., D. Karamanev, and A. Margaritis (2006) Kinetic modeling of the biodegradation of the aqueous p-xylene in the immobilized soil bioreactor. Biochem. Eng. J. 27: 204–211.
Liu, J., X. Jia, J. Wen, and Z. Zhou (2012) Substrate interactions and kinetics study of phenolic compounds biodegradation by Pseudomonas sp. cbp1-3. Biochem. Eng. J. 67: 156–166.
Texier, A. C. and J. Gomez (2007) Simultaneous nitrification and p-cresol oxidation in a nitrifying sequencing batch reactor. Water Res. 41: 315–322.
He, Z., H. Xiao, L. Tang, H. Min, and Z. Lu (2013) Biodegradation of di-n-butyl phthalate by a stable bacterial consortium, HD-1, enriched from activated sludge. Bioresour. Technol. 128: 526–532.
Adav, S. S., D. J. Lee, and J. Y. Lai (2009) Functional consortium from aerobic granules under high organic loading rates. Bioresour. Technol. 100: 3465–3470.
Patel, V., S. Jain, and D. Madamwar (2012) Naphthalene degradation by bacterial consortium (DV-AL) developed from Alang-Sosiya ship breaking yard, Gujarat, India. Bioresour. Technol. 107: 122–130.
Lee, D. J., K. Y. Show, and A. Wang (2013) Unconventional approaches to isolation and enrichment of functional microbial consortium: A review. Bioresour. Technol. 136: 697–706.
Polymenakou, P. N., C. A. Christakis, M. Mandalakis, and A. Oulas (2015) Pyrosequencing analysis of microbial communities reveals dominant cosmopolitan phylotypes in deep-sea sediments of the eastern Mediterranean Sea. Res. Microbiol. 166: 448–457.
Vedler, E., E. Heinaru, J. Jutkina, S. Viggor, T. Koressaar, M. Remm, and A. Heinaru (2013) Limnobacter spp. as newly detected phenol-degraders among Baltic Sea surface water bacteria characterised by comparative analysis of catabolic genes. Syst. Appl. Microbiol. 38: 525–532.
Marín, M. M., T. H. Smiths, J. B. van Beilen, and F. Rojo (2001) The alkane hydroxylase gene of Burkholderia cepacia RR10 is under catabolic repression control. J. Bacteriol. 183: 4202–4209.
Guo, C., Z. Dang, Y. Wong, and N. F. Tam (2010) Biodegradation ability and dioxgenase genes of PAH-degrading Sphingomonas and Mycobacterium strains isolated from mangrove sediments. Int. Biodeterior. Biodegrad. 64: 419–426.
Kawasaki, A. and M. A. Kertesz (2010) Hydrocarbon-degrading sphingomonads: Sphingomonas, sphingobium, novosphingobium, and sphingopyxis. Handb Hydrocarbon Lipid Microbiol. 1693–1705.
Kobayashi, T., Y. Murai, K. Tatsumi, and Y. Iimura (2009) Biodegradation of polycyclic aromatic hydrocarbons by Sphingomonas sp. enhanced by water-extractable organic matter from manure compost. Sci. Total Environ. 407: 5805–5810.
Adav, S. S., D. J. Lee, K. Y. Show, and J. H. Tay (2008) Aerobic granular sludge: Recent advances. Biotechnol. Adv. 26: 411–423.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Canul-Chan, M., Chable-Naal, J., Rojas-Herrera, R. et al. Hydrocarbon degradation capacity and population dynamics of a microbial consortium obtained using a sequencing batch reactor in the presence of molasses. Biotechnol Bioproc E 22, 170–177 (2017). https://doi.org/10.1007/s12257-016-0499-4
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12257-016-0499-4