Summary
Background
Acute lymphoblastic leukemia (ALL), characterized by overproduction and accumulation of immature lymphoid cells in bone marrow and peripheral blood, is the most common malignancy in children. NOTCH signaling is suggested to be a key event in hematological malignancies and appears to be a major oncogenic trigger in leukemia. Several studies on NOTCH target gene (HES‑1, p21 and c‑Myc) expression evaluated the correlation between these genes in AML (acute myeloid leukemia), but this relationship has not yet been clarified in ALL. Therefore, we aimed to study the expression of these genes in our Egyptian patients with ALL to obtain more information.
Patients and methods
RNA was extracted from peripheral blood mononuclear cells (PBMNCs) of 91 pediatric ALL patients (49 B-cell acute lymphoblastic leukemia [B-ALL] and 42 T-cell acute lymphoblastic leukemia [T-ALL]) and 52 healthy controls. The expression levels were determined by quantitative real-time polymerase chain reaction (qRT-PCR).
Results
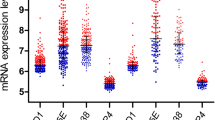
Median p21 and HES1 expressions were down regulated, while c‑Myc expression was up-regulated in B‑ALL cases (p < 0.001, p = 0.008, p < 0.001, respectively) and in T‑ALL cases (p = 0.049, p = 0.015, p < 0.001, respectively) when compared to the control group. Median HES1 expression was down regulated, in B‑ALL cases compared to T‑ALL cases (p = 0.002), while P21 and c‑Myc did not differ significantly between B‑ALL and T‑ALL cases.
Conclusion
P21 expression showed a significant positive correlation with HES1 expression and c‑Myc showed nonsignificant negative correlations with p21 and HES1, thus, suggesting that HES1 may affect ALL cells through the HES1–p21 pathway. Patients with over expressed c‑Myc had worse survival than patients with low expression which suggested it is a risk predictor.



Similar content being viewed by others
References
Maloney KW, Carroll WL, Carroll AJ, Devidas M, Borowitz MJ, Martin PL, et al. Down syndrome childhood acute lymphoblastic leukemia has a unique spectrum of sentinel cytogenetic lesions that influences treatment outcome: a report from the Children’s Oncology Group. Blood. 2010;116:1045–50. https://doi.org/10.3324/haematol.2010.024968.
Shalaby H, Ashaat A, El-Wahab A, El-Hamid M, El-Wakeel S. Bcl‑2 expression and chromosomal abnormalities in childhood acute lymphoblastic leukemia. Acad J Cancer Res. 2010;3:34–43.
Koch U, Radtke F. Notch and cancer: a double-edged sword. Cell Mol Life Sci. 2007;64:2746–62. https://doi.org/10.1007/s00018-007-7164-1.
Borggrefe T, Oswald F. The Notch signaling pathway: transcriptional regulation at Notch target genes. Cell Mol Life Sci. 2009;66:1631–46.
Qiao L, Wong BC. Role of Notch signaling in colorectal cancer. Carcinogenesis. 2009;30:1979–86. https://doi.org/10.1093/carcin/bgp236.
Iso T, Kedes L, Hamamori Y. HES and HERP families: multiple effectors of the Notch signaling pathway. J Cell Physiol. 2003;194:237–55. https://doi.org/10.1002/jcp.10208.
Grandori C, Cowley SM, James LP, Eisenman RN. The Myc/Max/Mad network and the transcriptional control of cell behavior. Annu Rev Cell Dev Biol. 2000;16:653–99. https://doi.org/10.1146/annurev.cellbio.16.1.653.
Levens DL. Reconstructing Myc. Genes Dev. 2003;17:1071–7. https://doi.org/10.1101/gad.1095203.
Felton-Edkins ZA, Kenneth NS, Brown TR, Daly NL, Gomez-Roman N, Grandori C, et al. Direct regulation of RNA polymerase III transcription by RB,p53 and c‑Myc. Cell Cycle. 2003;2:180–3.
Arabi A, Wu S, Ridderstråle K, Bierhoff H, Shiue C, Fatyol K, et al. c‑Myc associates with ribosomal DNA and activates RNA polymerase I transcription. Nat Cell Biol. 2005;7:303. https://doi.org/10.1038/ncb1225.
Grandori C, Gomez-Roman N, Felton-Edkins ZA, Ngouenet C, Galloway DA, Eisenman RN, et al. c‑Myc binds to human ribosomal DNA and stimulates transcription of rRNA genes by RNA polymerase I. Nat Cell Biol. 2005;7:311. https://doi.org/10.1038/ncb1224.
Grewal SS, Li L, Orian A, Eisenman RN, Edgar BA. Myc-dependent regulation of ribosomal RNA synthesis during Drosophila development. Nat Cell Biol. 2005;7:295. https://doi.org/10.1038/ncb1223.
Taniguchi T, Endo H, Chikatsu N, Uchimaru K, Asano S, Fujita T, et al. Expression of p21Cip1/Waf1/Sdi1 and p27Kip1cyclin-dependent kinase inhibitors during human hematopoiesis. Blood. 1999;93:4167–78.
El-Deiry WS, Harper JW, O’Connor PM, Velculescu VE, Canman CE, Jackman J, et al. WAF1/CIP1 is induced in p53-mediated G1 arrest and apoptosis. Cancer Res. 1994;54:1169–74.
Tian C, Tang Y, Wang T, Yu Y, Wang X, Wang Y, et al. HES1 is an independent prognostic factor for acute myeloid leukemia. OTT. 2015;8:899. https://doi.org/10.1007/s00277-015-2413-0.
Kannan S, Fang W, Song G, Mullighan CG, Hammitt R, McMurray J, et al. Notch/HES1-mediated PARP1 activation: a cell type–specific mechanism for tumor suppression. Blood. 2011;117:2891–900. https://doi.org/10.1182/blood-2009-12-253419.
Chen YQ, Cipriano SC, Arenkiel JM, Miller FR. Tumor suppression by p21WAF1. Cancer Res. 1995;55:4536–9.
Bae I, Fan S, Bhatia K, Kohn KW, Fornace AJ, O’Connor PM. Relationships between G1 arrest and stability of the p53 and p21Cip1/Waf1 proteins following γ‑irradiation of human lymphoma cells. Cancer Res. 1995;55:2387–93.
Polak J, Pekova S, Schwarz J, Kozak T, Haskovec C. Expression of cyclin-dependent kinase inhibitors in leukemia. Cas Lek Ceskych. 2003;142:25–8.
Watanabe M, Nakahata S, Hamasaki M, Saito Y, Kawano Y, Hidaka T, et al. Downregulation of CDKN1A in adult T‑cell leukemia/lymphoma despite overexpression of CDKN1A in human T‑lymphotropic virus 1‑infected cell lines. J Virol. 2010;84:6966–77. https://doi.org/10.1128/JVI.00073-10.
Scott SA, Kimura T, Dong W‑F, Ichinohasama R, Bergen S, Kerviche A, et al. Methylation status of cyclin-dependent kinase inhibitor genes within the transforming growth factor beta pathway in human T‑cell lymphoblastic lymphoma/leukemia. Leuk Res. 2004;28:1293–301.
Grabher C, von Boehmer H, Look AT. Notch 1 activation in the molecular pathogenesis of T‑cell acute lymphoblastic leukaemia. Nat Rev Cancer. 2006;6:347. https://doi.org/10.1038/nrc1880.
Weng AP, Millholland JM, Yashiro-Ohtani Y, Arcangeli ML, Lau A, Wai C, et al. c‑Myc is an important direct target of Notch1 in T‑cell acute lymphoblastic leukemia/lymphoma. Genes Dev. 2006;20:2096–109.
Palomero T, Lim WK, Odom DT, Sulis ML, Real PJ, Margolin A, et al. NOTCH1 directly regulates c‑MYC and activates a feed-forward-loop transcriptional network promoting leukemic cell growth. Proc Natl Acad Sci. 2006;103:18261–6. https://doi.org/10.1073/pnas.0606108103.
Gutierrez A, Sanda T, Ma W, Zhang J, Grebliunaite R, Dahlberg S, et al. Inactivation of LEF1 in T‑cell acute lymphoblastic leukemia. Blood. 2010;115:2845–51. https://doi.org/10.1182/blood-2009-07-234377.
Faderl S, O’Brien S, Pui CH, Stock W, Wetzler M, Hoelzer D, et al. Adult acute lymphoblastic leukemia: concepts and strategies. Cancer. 2010;116:1165–76. https://doi.org/10.1002/cncr.24862.
Malempati S, Tibbitts D, Cunningham M, Akkari Y, Olson S, Fan G, et al. Aberrant stabilization of c‑Myc protein in some lymphoblastic leukemias. Leukemia. 2006;20:1572.
Rice KL, Hormaeche I, Doulatov S, Flatow JM, Grimwade D, Mills KI, et al. Comprehensive genomic screens identify a role for PLZF-RARα as a positive regulator of cell proliferation via direct regulation of c‑MYC. Blood. 2009;114:5499–511. https://doi.org/10.1182/blood-2009-03-206524.
Acknowledgements
The authors like to thank the patients and healthy volunteers who participated in this study.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
M. Reyad, S. Abdel-Aziz, L.M. Saleh, S. El-Ghlban, I. El Tantawy El Sayed and H. Abdel-ghaffar declare that they have no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Reyad, M., Abdel-Aziz, S., Saleh, L.M. et al. The emerging role of NOTCH target genes in Egyptian childhood acute lymphoblastic leukemia. memo 14, 119–126 (2021). https://doi.org/10.1007/s12254-020-00665-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12254-020-00665-2