Abstract
Human respiratory syncytial virus (RSV) infection is the leading cause of lower respiratory tract illness (LRTI), and no vaccine against LRTI has proven to be safe and effective in infants. Our study assessed attenuated recombinant RSVs as vaccine candidates to prevent RSV infection in mice. The constructed recombinant plasmids harbored (5′ to 3′) a T7 promoter, hammerhead ribozyme, RSV Long strain antigenomic cDNA with cold-passaged (cp) mutations or cp combined with temperature-sensitive attenuated mutations from the A2 strain (A2cpts) or further combined with SH gene deletion (A2cptsΔSH), HDV ribozyme (δ), and a T7 terminator. These vectors were subsequently co-transfected with four helper plasmids encoding N, P, L, and M2-1 viral proteins into BHK/T7-9 cells, and the recovered viruses were then passaged in Vero cells. The rescued recombinant RSVs (rRSVs) were named rRSV-Long/A2cp, rRSV-Long/A2cpts, and rRSV-Long/A2cptsΔSH, respectively, and stably passaged in vitro, without reversion to wild type (wt) at sites containing introduced mutations or deletion. Although rRSV-Long/A2cpts and rRSV-Long/A2cptsΔSH displayed temperature-sensitive (ts) phenotype in vitro and in vivo, all rRSVs were significantly attenuated in vivo. Furthermore, BALB/c mice immunized with rRSVs produced Th1-biased immune response, resisted wtRSV infection, and were free from enhanced respiratory disease. We showed that the combination of ΔSH with attenuation (att) mutations of cpts contributed to improving att phenotype, efficacy, and gene stability of rRSV. By successfully introducing att mutations and SH gene deletion into the RSV Long parent and producing three rRSV strains, we have laid an important foundation for the development of RSV live attenuated vaccines.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Human respiratory syncytial virus (RSV) is the most common cause of pneumonia and the key factor in pneumonia-related deaths among children under five years of age. It is also known to cause deaths in the elderly and immunocompromised people (Falsey et al. 2005; Jorquera et al. 2016; Troeger et al. 2018; PERCH 2019). The earlier formaldehyde-inactivated RSV vaccine (FI-RSV) did not prevent RSV infection but resulted in enhanced respiratory disease (ERD) in the vaccinated children (Kapikian et al. 1969). Therefore, safety is set as the priority for the development of RSV vaccine for infants, and avoiding ERD as well as stimulating effective immune protection are still the main challenges of RSV vaccine research. One of the promising candidates is the RSV live attenuated vaccine (LAV), which has been successfully obtained by introducing mutations or/and deleting non-essential genes from the RSV genome (Connors et al. 1995; McFarland et al . 2018; McFarland et al. 2020a; McFarland et al. 2020b). Among the dispensable genomic regions are genes encoding small hydrophobic protein (SH), nonstructural proteins (NS), and M2 ORF protein 2 (M2-2) proteins. The deletions of NS1, NS2, or M2-2 are known to be promising strategies for obtaining intranasal RSV LAV against neonatal RSV infection (Luongo et al. 2013; McFarland et al. 2020b; Teng et al. 2020).
RSV LAV originating from the RSV Long parent strain has not yet been obtained; however, the RSV A2 parent has been widely used to construct RSV LAV candidates (Huang et al. 2009; Hu et al. 2014; Xu et al. 2018; Bouillier et al. 2019). RSV Long and A2 strains belong to the same GA1 clade and RSV group A. Moreover, their attachment glycoproteins (G) share 94% sequence identity (Johnson et al. 1987; Peret et al. 1998). Compared with the G protein, the fusion glycoprotein (F) is even more highly conserved among A and B RSV groups. A few RSV vaccine candidates have been developed by neutralizing antigenic F or G proteins from the RSV Long strain (Singh et al. 2007; Huang et al. 2009; Magro et al. 2012; Blanco et al. 2014). RSV LAV candidates from the RSV A2 parent have also been developed; however, different laboratory stocks of RSV parents differed in the level of attenuation (Lawlor et al. 2013; McFarland et al. 2018). Hence, we aimed to develop RSV LAV candidates from the RSV Long parent using a reverse genetics technology platform and determine whether their efficacy is comparable to that of RSV LAV from the RSV A2 strain.
The known examples of RSV LAV are mainly attenuated by point mutation, gene deletion, or by their combination. The original set of mutations included five cold-passaged (cp) mutations, and the temperature-sensitive (ts) point mutations identified by reverse genetics method (Crowe et al. 1994a, b, 1995). The replication ability of cpts-248/404 in vitro was weakened at 37 °C and the virus remained attenuated in infants attending the clinical trials. However, viral replication in the nasal cavity caused serious nasal congestion. Another RSV LAV (rA2cp248/404/1030ΔSH) was generated by SH gene deletion (ΔSH) (Bukreyev et al. 1997).
Here, we cloned plasmids containing RSV antigenomic cDNA from the wild-type RSV (wtRSV) Long strain harboring cp mutations, cpts mutations, or cptsΔSH mutations, and named them pBRB-RSV-rLong/A2cp, pBRB-RSV-rLong/A2cpts, and pBRB-RSV-rLong/A2cptsΔSH, respectively. Then, we co-transfected each of the constructed recombinant plasmids together with four helper plasmids encoding nucleoprotein (N), phosphoprotein (P), large protein (L), and M2-1 into BHK/T7-9 cells expressing T7 RNA polymerase, and the recovered viral particles were blind passaged in Vero cells. The successfully rescued recombinant RSVs (rRSVs), rRSV-Long/A2cp, rRSV-Long/A2cpts, and rRSV-Long/A2cptsΔSH, were analyzed for their genetic stability by sequencing and for their attenuation (att) phenotypes by measuring replication activity on the basis of growth curves. The strains were also assessed for temperature sensitivity by measuring viral titers at different temperatures. Finally, BALB/c mice were intranasally inoculated with each rRSV and subsequently challenged with wtRSV. The immunogenicity, safety, and efficacy of these rRSVs were evaluated on the basis of serum antibody levels, neutralizing antibody titers, lung pathology, and viral replication activity in the upper and lower respiratory tracts.
Materials and Methods
Cells, Viruses, and Plasmids
HEp-2 cells and Vero cells (ATCC, Rockefeller, MD, USA) were grown in Dulbecco's Modified Eagle Medium (DMEM) (Gibco BRL, Gaithersburg, USA) containing 2 mmol/L l-glutamine and 10% fetal bovine serum (FBS, Gibco, Australia). BHK/T7-9 cells were kindly provided by Professor W. Y. Zhu (CDC, Beijing, China) and maintained in DMEM with 10% tryptose phosphate broth (Sigma, Darmstadt, Germany), 5% FBS, 2 mmol/L l-glutamine (Amresco, Solon, USA) and antibiotics (100 IU/mL penicillin, 10 μg/mL streptomycin, 0.25 μg/mL amphotericin B, and 600 ng/mL hygromycin, Sigma). wtRSVs of subgroup A strain Long and subgroup B (WV VR1400, ATCC) were propagated using HEp-2 cells cultured in DMEM supplemented with 2% FBS, l-glutamine (2 mmol/L), antibiotics (40 IU/mL penicillin G, 100 μg/mL streptomycin), and 0.2% sodium bicarbonate. The helper plasmids, pCITE-N, pCITE-P, pCITE-L, and pCITE-M2-1, encoding N, P, L, and M2-1 proteins, respectively, were gifts from Dr. Marie-Anne Rameix-Welti (Unité de Virologie et Immunologie Moléculaires, Université Paris-Saclay, Paris, France).
Rescue and Identification of Recombinant RSVs
The construction and recovery of rRSV-Long/A2cp, rRSV-Long/A2cpts, and rRSV-Long/A2cptsΔSH were performed as reported previously (Xu et al. 2018). Briefly, to construct antigenomic cDNA clones, each of the mentioned mutations and deletion was introduced into the full-length wtRSV cDNA. Specifically, the deletion included the entire SH gene or the majority of the downstream noncoding region of the SH gene, as well as silent nucleotide (nt) substitutions in the last three codons and the termination codon of the SH ORF, leaving the gene end signal intact (Bukreyev et al. 2001). The resulting plasmids, containing antigenomic cDNAs flanked by T7 promoter-hammerhead ribozyme in the 5′ end and by HDV ribozyme-T7 terminator in the 3′ end (Fig. 1A), were named pBRB-RSV-rLong/A2cp, pBRB-RSV-rLong/A2cpts, and pBRB-RSV-rLong/A2cptsΔSH, respectively. For rescue of rRSVs, BHK/T7-9 cells were co-transfected with a plasmid harboring rRSV antigenomic cDNA and four helper plasmids (pCITE-N, pCITE-P, pCITE-L, and pCITE-M2-1) using Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA). The transfection mixtures were prepared as follows: rRSV cDNA plasmid 1.25 μg, pCITE-N 1 μg, pCITE-P 1 μg, pCITE-L 0.5 μg, pCITE-M2-1 0.25 μg. The mixtures of DNA-lipo-OptiMEM (Gibco BRL, Gaithersburg, MD, USA) were added into cells after a 20-min incubation. The cells were incubated at 33 °C in a 5% CO2 incubator. For the blind passage, Vero cells were incubated with 400 μL of the suspension from the harvested co-transfected cells at 33 °C for 2 h. Then, the infected Vero cells were incubated with DMEM containing 2% FBS at 33 °C and expanded or serially passaged. They were monitored by immunoplaque assay and real-time quantitative PCR (RT-qPCR) as described previously (van Elden et al. 2003; Fu et al. 2013; Jiao et al. 2017). For the immunoplaque assay, serially diluted RSV samples were grown to 80% confluency of HEp-2 cells in a 96-well plate in triplicate for 1 h at 37 °C, and then DMEM containing 0.9% methyl cellulose (Sigma) was added to the wells. Subsequently, the media were discarded and the cells were rinsed with DMEM without FBS. After 3 days of incubation at 37 °C under 5% CO2, the cell monolayer was fixed with 95% cold alcohol, and viral replication activity could be assessed using an goat anti-RSV antibody (Millipore, Billerica, MA, USA) incubated with horseradish peroxidase rabbit anti-goat IgG (Santa Cruz Biotechnology, Santa Cruz, CA, USA), and visualized after adding TMB (Promega, Madison, WI, USA). RSV titers were expressed as plaque-forming units per mL (pfu/mL). For RT-qPCR, RNA samples from virus-infected cells were extracted using Trizol reagent (Invitrogen) according to the manufacturer’s instructions, and quantified on the basis of the obtained OD value. The reverse transcription was completed with the GoScript™ reverse transcription system (Promega). Briefly, 1.5 μg RNA samples were incubated with oligo dT for 5 min at 70 °C and then placed for 5 min on ice. Following the addition of 15 μL of RT reaction mix, the samples were placed for 5 min at 25 °C, 1 h at 42 °C, and 15 min at 70 °C, and then stored at 4 °C. Subsequently, qPCR was performed by using a SYBR green probe (Tiangen Biotech, Beijing, China). The primers for the RSV N gene were as follows: forward primer, 5′-AGATCAACTTCTGTCATCCAGCAA-3′ and reverse primer, 5′-GCACATCATAATTAGGAGTATCAAT-3′. The thermal cycling conditions included 15 min at 95 °C, followed by 45 cycles of 15 s at 9 °C and 1 min at 60 °C. The specificity of the obtained qPCR products was verified by melting point analysis in the range from 45 to 95 °C.
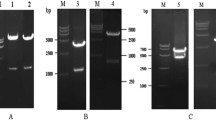
Schematic diagram illustrating our strategy for the identification of recombinant human respiratory syncytial viruses (rRSVs). A Schematic diagram of rRSV genomes. The five missense mutations Val-267-Ile/N, Glu-218-Ala/F, Thr-523-Ile/F, Cys-319-Tyr/L, and His-1690-Tyr/L are independent attenuating genetic elements of cold-passaged (cp) mutation; Gln-831-Leu/248 and T-7605-C/404 are two independent temperature-sensitive (ts) mutations; ΔSH is a complete deletion of the SH gene. T7 Pro: T7 promoter, T7 Ter: T7 terminator, Le: Lead, Tr: Trail. B Identification of replication of the rescued rRSVs by immunoplaque assay. Vero cells were inoculated with suspensions from cells co-transfected with four helper plasmids (Mock), and with pBRB-RSV-rLong/A2cp, pBRB-RSV-rLong/A2cpts, or pBRB-RSV-rLong/A2cptsΔSH, respectively.
Preparation of rRSVs, wtRSV, and FI-RSV
The preparation and purification of wtRSV and rRSVs were performed in line with a previously described method (Kohlmann et al. 2009; Kwanten 2013; Jiao et al. 2017). Briefly, the viral samples were inoculated onto HEp-2 cells with 80% confluency and incubated for 3 days at 37 °C under 5% CO2. After the formation of syncytia, the cells were scraped off and centrifuged at 3000 rpm for 10 min at 4 °C, and the supernatants were collected. To further release intracellular viral particles, the cell pellet was exposed to three cycles of swift freezing/thawing (the cell pellet was immersed in liquid nitrogen for 10 s and then thawed using running tap water). The frozen/thawed pellet was centrifuged for 10 min at 3000 rpm and the supernatants were collected. The pooled supernatants were run through a 0.45 μm sterile filter (Merck Millipore, Co., Cork, Ireland), and purified by ultracentrifugation on a sucrose cushion gradient (10% sucrose, Sigma) at 17,000 rpm in a P28S rotor (Hitachi, Tokyo, Japan) for 2 h at 4 °C. The pellet was suspended in 300 μL 10% sucrose, divided into aliquots, and stored at − 80 °C. The infectivity of the RSV was assessed using an immunoplaque assay as described above.
FI-RSV was prepared according to a previously described method (Kim et al. 1969). Briefly, RSV-infected cell lysates were clarified by centrifugation for 15 min at 550 ×g. Then, the clarified cell lysates were incubated with 37% formalin (Sigma) for 3 days at 36 °C in the proportion of 1:4000. The mixture was then pelleted by ultracentrifugation at 17,000 rpm in a P28S rotor for 1 h at 4 °C. The resulting pellet was resuspended in 1/25 of the original volume in serum-free DMEM and assayed for protein concentration using a BCA protein assay kit (Thermo Fisher Scientific Inc., Waltham, MA, USA).
In vitro Characterization of rRSVs
The above mentioned immunoplaque assay and RT-qPCR were used to evaluate the replication activity and confirm the successful rescue of the rRSVs since passage 1 (P1) in Vero cells. Simultaneously, a GoScript™ reverse transcription (RT)-PCR kit was used to assess the genetic stability of the rescued rRSVs by amplifying fragments of viral DNA harboring the mutated nucleotides contributing to the att phenotype. The primers used for RT-PCR are shown in Supplementary Table S1. These characteristic DNA fragments were amplified for each rRSV sample from every odd-numbered passage from P1 to P9 and subsequently sequenced to monitor the possible reversion of individual point mutations.
The rRSV growth kinetics was also assayed using a previously described method (Collins and Bermingham 1999; Schickli et al. 2012). Briefly, HEp-2 cells were infected with wtRSV or rRSVs at a multiplicity of infection (MOI) of 0.1 in triplicate and incubated at 32 °C. Cells and supernatants were harvested at 24-h intervals post-infection, and viral titers were determined by immunoplaque assay as mentioned above.
The temperature-sensitive phenotype of rRSVs in vitro was evaluated by determining the efficiency of plaque formation at various temperatures, through a modified method from a previous report (Crowe et al. 1993). Briefly, a HEp-2 cell monolayer, grown in 24-well plates, was inoculated with tenfold serial dilutions of wtRSV and rRSV cultures, incubated for 5 days at 32 °C, 35 °C, 36 °C, 37 °C, 38 °C, 39 °C, and 40 °C, and then assayed for infectivity at each corresponding temperature using the immunoplaque assay.
The temperature stress test was performed as reported previously (Collins and Bermingham 1999; Schickli et al. 2012). Briefly, rRSVs were passaged at elevated non-permissive temperatures, twice at 37 °C, twice at 39 °C, and once at 40 °C. Next, HEp-2 cells were inoculated with rRSVs at 0.1 MOI in a 96-well plate. Following a 1-h incubation at 33 °C, the inoculum was collected by pipetting, and cells were supplied with 200 μL/well of DMEM with 2% FBS and incubated at 37 °C. Following a 5-day incubation at 37 °C, 100 μL of the media from each of the infected wells were transferred to an uninfected 96-well plate containing HEp-2 cells, in duplicate. After a 1-h incubation, the media were removed by pipetting and the cells were supplied with 200 μL/well of fresh medium. The plates were then incubated for 5 days at 37 °C for the second passage. The virus was similarly transferred and incubated at 39 °C to generate the third and fourth passages. The last passage was performed at 40 °C. For each passage, one of the duplicate HEp-2 plates was immunostained to assess RSV infectivity.
Replication of rRSVs in the Upper and Lower Respiratory Tracts of Mice
Six-week-old female BALB/c mice (Charles River Laboratories, Beijing, China) were housed in five groups under pathogen-free conditions in microisolator cages at the animal quarters of Tsinghua University Medical Center (Beijing, China). The replication of rRSVs was evaluated in the upper and lower respiratory tracts of mice as previously described (Whitehead et al. 1998a). Briefly, mice were infected via intranasal (i.n.) route by rRSVs or wtRSV (at 1 × 106 pfu/mouse) and sacrificed by CO2 inhalation for washes of nasal and lung tissues on days 2, 4, 6, or 8 post-infection. Viral titers in the harvested washes of nasal and lung tissues were determined using the immunoplaque assay and RT-qPCR as mentioned above. For RT-qPCR, viral RNA extracts from nasal and lung washes were obtained using Trizol reagent according to the manufacturer's instructions, and all the remaining RT-qPCR procedures were repeated as described previously.
Nasal washes were collected as previously reported (Rostad et al. 2016). Briefly, the jaws and the head of each sacrificed mouse were disarticulated and sequentially removed. Next, 1 mL of Iscove's Modified Dulbecco's Medium (IMDM) (Gibco, Paisley, Scotland, UK) containing 15% glycerin mixed with 2% FBS-MEM (1:1, vol/vol) was pushed through each naris (total of 2 mL). All nasal washes were collected and stored at − 80 °C.
The lungs were harvested from mice in each group as previously reported (Fu et al. 2014), weighed, placed in phosphate-buffered saline (PBS) containing 1% bovine serum albumin (100 μL/0.1 g lung), and homogenized with a glass tissue grinder. The lung homogenates were centrifuged at 12,000 ×g for 10 min and stored at − 80 °C.
Animal Vaccination and Challenge
BALB/c mice were randomly grouped (five mice/group), lightly anesthetized with avertin, and then subjected to either i.n. immunization with rRSVs (1 × 106 pfu/mouse) or wtRSV (1 × 106 pfu/mouse) or intramuscular (i.m.) immunization with FI-RSV (1.875 μg/mouse) in 50 μL of PBS with 10% sucrose. Alternatively, the mice were subjected to i.m. injection containing only 50 μL PBS with 10% sucrose and were labeled as the negative control group. For the challenge experiments, the immunized or negative control BALB/c mice were further lightly anesthetized and infected with 1 × 106 pfu of wtRSV at day 28 after vaccination.
Evaluation of RSV-Specific Serum IgG Responses by ELISA
Blood samples were collected as previously reported (Fu et al. 2014). Briefly, the samples were collected from the retro-orbital plexus of mice with a capillary tube. After centrifugation at 6000 ×g for 15 min, the obtained sera were stored at 4 °C. RSV-specific serum antibody responses (Jiao et al. 2017; Ma et al. 2018) were analyzed by adding tenfold dilutions of serum samples to the RSV-coated plates, which were then incubated for 1 h at 37 °C. Following three washes with PBS, the plates were incubated with HRP-conjugated anti-mouse IgG, IgG1, or IgG2 antibodies (1:5000 dilution, Santa Cruz Biotechnology) for 1 h, developed with 100 μL of TMB substrate solution (Sigma), stopped with 50 μL of 2 mol/L H2SO4, and analyzed at 450 nm using an ELISA plate reader (Tecan, Grodig, Austria). The titers of anti-RSV antibodies were expressed as the reciprocal of the maximal dilution of serum giving an absorbance reading greater than 0.2 absorbance units and being two-fold above the absorbance obtained for the negative control group.
RSV-Specific Neutralizing Antibody Assay
To analyze RSV-specific neutralizing antibody titer (Jiao et al. 2017), the samples of sera were heat-inactivated at 56 °C for 30 min and prepared by two-fold serial dilution in separate virus diluents. Fifty pfu of RSV virus suspensions (subgroup A wtRSV Long or subgroup B (WV VR1400)) were incubated with serially diluted samples for 1 h at 37 °C. Then, the neutralizing antibody titers were determined by immunoplaque assay as described above. Neutralization titers were expressed as the reciprocal of the dilution of serum giving a 50% reduction in the number of pfu in control wells.
Pulmonary Histology of RSV-Infected Mice
Histological analyses of lung tissues following RSV challenge were performed as described previously (Jiao et al. 2017). After mice were sacrificed by CO2 inhalation, the lungs were fixed via 4% formalin infusion through the trachea. The solution was removed and the lungs were subsequently immersed in 4% formalin for 24 h, embedded in paraffin, sectioned, and stained with hematoxylin and eosin (H&E). Ten sections of 100 fields per mouse were examined. Sections from each mouse were blindly scored for the degree of pulmonary inflammatory changes including peribronchiolitis, perivasculitis, alveolitis, and interstitial pneumonitis. They were graded as follows: 0, clear; 1, slight; 2, moderate; 3, abundant; and 4, severe.
The eosinophilia in lung tissues of RSV-challenged mice was determined as previously described (Jiao et al. 2017). Briefly, the formalin-fixed, paraffin-embedded lung tissue sections were stained with specific stain kits (ZSGB-BIO, Beijing, China), and interpreted by assessment of both staining distribution and intensity with Image-Pro Plus software (Media Cybernetics, Washington, D.C., USA).
Statistical Analyses
Statistical analyses of data were performed using GraphPad Prism 5 software (GraphPad Software, La Jolla, CA, USA). Differences were evaluated using independent, two-sided Student’s t test. Results characterized by P < 0.05 were considered statistically significant.
Results
Construction and Identification of rRSVs
The plasmids encoding rRSV-Long/A2cp, rRSV-Long/A2cpts, and rRSV-Long/A2cptsΔSH were obtained by a stepwise assembly of the synthesized cDNA segments. The locations of cp and ts mutations are shown in Fig. 1A. The full-length antigenomic cDNAs of pBRB-RSV-rLong/A2cp and pBRB-RSV-rLong/A2cpts were both expected to be 18,815 bp in size, while that of pBRB-RSV-rLong/A2cptsΔSH was expected to be 18,485 bp in size. The corresponding lengths were confirmed by DNA sequencing (data not shown).
For the recovery of rRSVs, the plasmid containing RSV antigenomic cDNA was co-transfected together with four plasmids encoding helper proteins to BHK/T7-9 cells, and the recovered rRSVs were subsequently blindly passaged in Vero cells. The rescued rRSVs were identified by immunoplaque assay as shown in Fig. 1B and by RT-PCR (data not shown). These results demonstrated that we successfully rescued the rRSVs bearing the anticipated cp, cpts, and cptsΔSH mutations.
In vitro Characterization of rRSVs
The titers of rRSV-Long/A2cp increased rapidly during the initial passages from passage 1 (P1) to P3 (P < 0.01) and remained constant after P3 as shown by immunoplaque assay and RT-qPCR. In contrast, the titers of rRSV-Long/A2cpts and rRSV-Long/A2cptsΔSH increased steadily until P4 (P < 0.01), and fluctuated marginally after P4 (Fig. 2A, 2B). The sequencing of the DNA fragments enclosing the individual rRSV point mutations determined the genetic stability of the rescued viruses. All the introduced att mutations were stable, and no reversion was detected. The sequencing results for passages from P1 to P9 are shown in Supplementary Table S2. To further characterize rRSVs, the growth kinetics of rRSVs and wtRSV in HEp-2 cells at 33 °C were assessed and compared between the two groups. The viral titers began to increase from 24 h post-inoculation and ultimately plateaued at 72 h post-inoculation for all the viruses. wtRSV achieved the titer of 4.5 × 107 pfu/mL, rRSV-Long/A2cp 2.8 × 106 pfu/mL, rRSV-Long/A2cpts 2.2 × 105 pfu/mL, and rRSV-Long/A2cptsΔSH 1.1 × 105 pfu/mL, as shown in Fig. 2C. All the three rRSVs exhibited approximately 10- or 100-fold reduced growth kinetics compared with that of the parent strain. Altogether, all rRSVs bearing the att mutations were constructed successfully, showed a markedly attenuated phenotype in vitro, and their proliferation rates were significantly lower when compared with that of the wtRSV parent.
In vitro characterization of recombinant human respiratory syncytial viruses (rRSVs). The replication titers during serial passages of rRSVs were monitored by immunoplaque assay (A) and by RT-qPCR (B) since passage 1 (P1). The growth curves for rRSVs and wild-type RSV (wtRSV) were established and compared (C). Each virus was harvested every other 24 h post-infection and titers were assayed by immunoplaque assay. All results are representative of three independent experiments. Data are shown as mean ± SD. *P < 0.05, **P < 0.01.
Analysis of ts Mutation Phenotype of rRSVs In Vivo and In Vitro
The ts phenotype of the rRSVs was examined by determining the efficiency of plaque formation by inoculation of tenfold viral dilutions at various temperatures in HEp-2 cells placed in TC24-well plates and comparing the results for wtRSV and rRSVs. The plates were incubated for 5 days at the specified temperatures in CO2 incubators calibrated to ± 1 °C and the average viral titers were measured by immunoplaque assay in HEp-2 cells at the corresponding temperatures (Table 1). Both rRSV-Long/A2cpts and rRSV-Long/A2cptsΔSH exhibited reduced viral titers by more than 2log10 at 37 °C compared to the permissive temperature of 32 °C and were therefore considered ts at ≥ 37 °C. As expected, the wtRSV as well as the precursor virus rRSV-Long/A2cp were not ts and showed no statistical titer reduction from 32 to 40 °C.
We also intranasally infected mice with 1 × 106 pfu of rRSVs and measured viral titers in samples obtained from nasal washes and lung tissue homogenates on days 2, 4, 6, and 8 post-infection by using immunoplaque assay and RT-qPCR (Fig. 3A–3D). The results showed that the titers of all three rRSVs in the nasal wash specimens at day 4 were 0.52–1.06 log10 total pfu, lower than that of wtRSV (P < 0.01–P < 0.001), and the titers in the lung tissues at day 4 and day 6 were 0.51–2.51 and 1.55–3.99 log10 pfu/g tissue, lower than the respective ones of wtRSV (P < 0.001). The number of RNA copies of the three rRSVs in the nasal wash specimens at day 4 was 0.32–0.88 log10 RSV copies/μg RNA, lower than that of wtRSV (P < 0.01–P < 0.001); in addition, in the lung tissues at days 4 and 6 the three rRSVs exhibited 1.00–2.55 and 1.20–3.74 log10 RSV copies/μg RNA, respectively, lower than the corresponding number of RNA copies of wtRSV (P < 0.001). These results are consistent with the corresponding immunoplaque test results. Among the three rRSVs, the two rRSVs possessing ts mutations displayed a higher level of attenuation than rRSV-Long/A2cp in the lung tissues (P < 0.001).
The attenuation (att) phenotype of recombinant human respiratory syncytial viruses (rRSVs) assayed in intranasally infected BALB/c mice. Viral titers in samples obtained from nasal washes were analyzed by immunoplaque test (A) and RT-qPCR (B); viral titers in lung tissues were analyzed by immunoplaque test (C) and RT-qPCR (D). Data are shown as mean ± SD. **P < 0.01, ***P < 0.001. LOD: limit of detection.
We also found that the replication titers of each rRSV bearing ts mutations were significantly higher in the samples obtained from nasal washes than in those obtained from lung tissue homogenates (P < 0.05 or P < 0.01), as shown in Table 1. Consistent with their in vitro ts phenotypes, the two rRSVs harboring ts mutations displayed shut-off temperatures equal to 37 °C and are therefore more competent to multiply in the nasal cavity environment, characterized by lower temperatures (Table 1).
To determine both the genetic and phenotypic stability of rRSVs, rRSV-Long/A2cpts was passaged in vitro at non-permissive temperatures to induce mutation, in line with the classical theory of the survival of the fittest. Briefly, rRSV-Long/A2cpts was serially passaged twice at 37 °C, and then subjected to two further passages at 39 °C and one passage at 40 °C (Fig. 4). After each passage, one of the duplicate plates was immunostained with anti-RSV antibodies. Initially, after the expansion at 37 °C, all the wells exhibited positive RSV immunostaining. After P4 at 39 °C, more than 80% of the wells had positive RSV immunostaining, suggesting the presence of the temperature-sensitive intermediate (tsi) viruses at this temperature. At 40 °C, the control wells containing wtRSV all had positive immunostaining. In sharp contrast, only 20%–30% of the wells containing rRSV-Long/A2cpts had positive RSV immunostaining. To investigate the nt changes at the ts markers, we analyzed the sequence of 1–2 kb cDNA fragments spanning from the start of the M2 gene through the L gene. RT-PCR was performed on five randomly chosen potential tsi revertants from RSV-positive wells to detect nt changes at the 248 and 404 ts sites of the L and M2 genes by sequence analysis. The characteristics of the biologically derived tsi viruses from the passaging of rRSV-Long/A2cpts at 39 °C are listed in Supplementary Table S3. The tsi strain were characterized by nt changes causing the reversion to the wt nt or amino acid at the 248 or the 404 ts markers and the partial loss of temperature sensitivity.
Passaging of rRSV-Long/A2cpts at 37 °C and 39 °C. To determine the frequency of intermediate temperature sensitive (tsi) viruses, two plates of HEp-2 cells were inoculated at multiplicity of infection (MOI) = 0.1 and supplied with 200 μL of medium. At 5 days post-infection new uninfected HEp-2 cells in 96-well plates were inoculated with 100 μL/well of supernatant from the previous virus passage. Following a 1-h incubation, the inoculum was removed and the cells were fed with 200 μL of medium. Blind passaging was performed in duplicate and at progressively higher temperatures. For each passage, one plate was immunostained and a duplicate plate was used to seed the next passage. Six wells of HEp-2 cells infected with wild-type RSV (wtRSV) were used as a positive control.
Characteristics of RSV-Specific Immune Responses and Neutralizing Antibody Responses
To study the immunogenicity of the rRSVs, mice were immunized intranasally with 1 × 106 pfu of rRSVs or wtRSV. Mice inoculated with PBS were used as negative control. Serum samples were collected 21 days after immunization, and the levels of RSV-specific serum IgG were detected by ELISA (Fig. 5A). The mice in both immunized groups produced significant RSV-specific IgG responses compared to the negative control (P < 0.001), but there was no difference between the IgG levels induced by the three rRSVs (P > 0.05). Compared with wtRSV, similar immunogenicity was observed for rRSV-Long/A2cp and rRSV-Long/A2cptsΔSH (P > 0.05). However, rRSV-Long/A2cpts was characterized by a slightly lower level of immunogenicity than that of wtRSV (P < 0.05).
RSV-specific immune responses and neutralizing antibody responses following infection with recombinant human respiratory syncytial viruses (rRSVs) and wild-type RSV (wtRSV). BALB/c mice were immunized with either rRSV-Long/A2cp, rRSV-Long/A2cpts, rRSV-Long/A2cptsΔSH, or wtRSV at 1 × 106 pfu/mouse or PBS (50 μL/mouse), via intranasal (i.n.) route, or FI-RSV at 1.875 μg/mouse via intramuscular (i.m.) route at day 0. Serum antibody titers against RSV were examined by ELISA (A), and the levels of induced serum neutralizing antibodies were assessed by immunoplaque assay (B) at day 21 post-immunization. The results for the neutralizing antibodies were expressed as the reciprocal of the highest serum dilution fold providing 50% inhibition of plaque formation. The induced isotype of RSV-specific IgG levels in BALB/c mice after immunization with rRSVs were assessed by ELISA (C), and ratios of IgG2a and IgG1 were calculated (D). nc: negative control group. Data are shown as mean ± SD. ns: no significance, *P < 0.05, **P < 0.01, ***P < 0.001.
The cross-protection against subgroup A wtRSV Long and subgroup B (WV VR1400) infection was also detected and the neutralizing antibody level was measured (Fig. 5B). Similar to the IgG responses, no significant difference in the levels of neutralizing antibodies was induced by immunization with the three rRSVs (P > 0.05). Analogically, when compared with wtRSV, similar neutralizing antibody responses and cross-protective effects were induced by rRSV-Long/A2cp and rRSV-Long/A2cptsΔSH (P > 0.05). However, rRSV-Long/A2cpts was characterized by a slightly lower response level (P < 0.05).
To determine the type of immune response induced in mice by rRSV immunization, the titers of IgG2a and IgG1, RSV-specific IgG subtypes, were examined, and the IgG2a/IgG1 ratio was calculated. The results showed that both rRSV and wtRSV immunization elicited a Th1-biased immune response, different from the Th2-biased immune response induced by intramuscular immunization with FI-RSV (Fig. 5C, 5D). In addition, the FI-RSV immunized mice also showed a significantly reduced neutralizing antibody response compared with mice in the wtRSV group (P < 0.01). However, the IgG response was similar between both FI-RSV and wtRSV immunization groups.
The Efficacy of Intranasal Vaccination with rRSVs
To evaluate the efficacy of RSV vaccine candidates, changes in body weight and lung viral titers were examined in vaccinated mice after RSV challenge. The mice were challenged with wtRSV 28 days after rRSV immunization, and their body weight was monitored for 5 consecutive days (Fig. 6A). Weight loss in challenged mice immunized by wtRSV and the three rRSVs was significantly reduced from day 2 to day 5 compared to that of FI-RSV-immunized mice (P < 0.05 or P < 0.001). On the other hand, mice immunized by either rRSV-Long/A2cp or rRSV-Long/A2cptsΔSH did not display significant differences in weight loss compared with that of wtRSV (P > 0.05), but a significant difference was observed between mice immunized with rRSV-Long/A2cpts and wtRSV on day 3 and day 4 after the challenge (P < 0.05). Moreover, the immunization with rRSV-Long/A2cpts resulted in weight changes different from those induced by rRSV-Long/A2cp or rRSV-Long/A2cptsΔSH at day 3 after challenge (P < 0.05, data not shown). The slightly decreased neutralizing antibody response observed in mice after immunization with rRSV-Long/A2cpts when compared to other rRSVs, mentioned above, may be an explanation for this phenomenon. Altogether, rRSV-Long/A2cp and rRSV-Long/A2cptsΔSH exhibited the best efficacy among the three rRSVs, which was reflected by weight changes after wtRSV challenge.
Induction of protective immunity against human respiratory syncytial virus (RSV) infection in the vaccinated BALB/c mice. After the immunized BALB/c mice were challenged with wild-type RSV (wtRSV, 1 × 106 pfu/mouse) at day 28 post-immunization, the body weight changes were monitored daily from day 1 to day 5 following viral challenge (A). Lung homogenates were collected on day 5 post-challenge, and lung viral loads were determined by immunoplaque assay (B) and RT-qPCR (C). nc: negative control group. Data are shown as mean ± SD. ns: no significance, *P < 0.05, **P < 0.01, ***P < 0.001.
On day 5 after the challenge, the lung tissues were collected from the examined mice. Lung viral titers were analyzed by immunoplaque assay and RT-qPCR (Fig. 6B, C). Immunization with wtRSV and each rRSV resulted in decreased lung viral titers when compared to those observed in mice immunized with FI-RSV and mice from the negative control group (P < 0.001). On the other hand, there were no significant differences in lung viral titers among mice immunized with the three rRSVs (P > 0.05). However, the lung viral loads were significantly higher in mice from the rRSV-Long/A2cp and rRSV-Long/A2cpts groups than in the wtRSV-immunized group (P < 0.05). Further, no observable differences existed between the lung viral titers of mice immunized with rRSV-Long/A2cptsΔSH and wtRSV after wtRSV challenge (P > 0.05). These results showed that all rRSV-immunized mice were protected against RSV infection; however, rRSV-Long/A2cptsΔSH provided the best protection.
Absence of Pulmonary Pathology and Eosinophilia in rRSV-Immunized Mice Following RSV Infection
Since the FI-RSV vaccine causes ERD in immunized children and animals, the pathological examination of lungs in primed mice after RSV challenge is an important safety index for the RSV vaccine candidates. To this purpose, we collected the mouse lung tissues for histological sections 5 days after the challenge, performed H&E staining, and evaluated the pulmonary pathology under a microscope (Fig. 7A). The alveolar walls in mice immunized with FI-RSV were thickened, and alveolar cavities were compressed to form lung parenchyma. There was also a large number of inflammatory cells infiltrating around the blood vessels and bronchi. In negative control mice, the alveolar walls appeared thickened, the alveolar septa collapsed resulting in cavitation, and inflammatory cell infiltration was present around the blood vessels and bronchi. In the wtRSV- and rRSV-Long/A2cpts-immunized mice, thickening alveolar walls, cavitation, and increased levels of inflammatory cells were also observed in the lung tissues, albeit to a less extent. On the other hand, the immunization of mice with rRSV-Long/A2cp and rRSV-Long/A2cptsΔSH resulted in clear as well as less destroyed alveoli, and only a few inflammatory cells accumulated around the bronchi.
Absence of pulmonary histopathology and eosinophilia in mice vaccinated with recombinant human respiratory syncytial viruses (rRSVs) following RSV challenge. After the immunized BALB/c mice were challenged with wild-type RSV (wtRSV, 1 × 106 pfu/mouse) at day 28 post-immunization, lung tissues were collected at day 5 post-challenge and pulmonary histopathology was analyzed by hematoxylin and eosin staining (H&E) (A). The H&E-stained lung sections from each mouse were scored for inflammation, including peribronchiolar inflammation (B), perivascular inflammation (C), interstitial pneumonia (D), and alveolitis (E). Distribution and intensity of eosinophils were measured and analyzed using specific stains and Image-Pro Plus (F). IOD: integrated optical density. nc: negative control group. Data are shown as mean ± SD. *P < 0.05, **P < 0.01, ***P < 0.001.
Lung sections from all mice were scored for inflammation around airways, blood vessels, and interstitial and alveolar spaces as described in the Materials and Methods (Fig. 7B–7E). A considerably severe (score around 4) lung histopathology was observed in mice vaccinated with FI-RSV while a less severe lung histopathology (score around 2) was characteristic for mice in the negative control group. The least severe lung histopathology (score range, 1 to 2) was observed in at least two mice in each rRSV-vaccinated group (rRSV-Long/A2cp and rRSV-Long/A2cptsΔSH) compared to the FI-RSV vaccination group (P < 0.001) and the negative control group (either P < 0.05, P < 0.01, or P < 0.001). These results showed that vaccination with two of the three rRSVs resulted in protective immunity without causing any obvious signs of ERD, while the documented FI-RSV immunization-triggered ERD after viral challenge was confirmed. Altogether, the observed pulmonary pathology exhibited similar trends to those of the relative body weight loss among the experimental groups.
In the lungs of mice immunized with FI-RSV we also observed the infiltration of eosinophils, unlike in the lungs of mice from the rRSVs and wtRSV vaccination groups (P < 0.001) (Fig. 7F). In particular, the lungs of mice immunized with rRSV-Long/A2cptsΔSH displayed the least number of eosinophils in comparison with the wtRSV vaccination group and the negative control group (P < 0.05; P < 0.01). This result is consistent with the histopathological observations, and indicates decreased inflammation owing to a better protection against RSV infection through immunization with rRSV-Long/A2cptsΔSH.
Discussion
To our knowledge, this is the first report on the successful construction and rescue of rRSVs obtained from a parental strain other than A2 (parental strain Long) and bearing cp, cpts, or cptsΔSH mutations. Most of the cDNA-derived rRSVs are based on the cpRSV and cptsRSV parents, which are both biologically derived live attenuated viruses from the RSV A2 strain. Although differences exist in the genomic makeup as well as the encoded proteins (such as G protein) of wtRSV Long and RSV A2, both strains are classified as group A and genotype GA1 RSVs (Peret et al. 1998). Therefore, we aimed to examine the characteristics of RSV LAV candidates obtained from cDNA of the wtRSV Long parent rather than the biologically derived one, and used the rRSV-Long/A2cp parent, a counterpart of the biologically derived cpRSV.
The att phenotype and genetic stability are the critical features of live attenuated RSV vaccine candidates. We found that rRSV-Long/A2cp is non-ts, similar to the biologically derived cpRSV and rRSVA2cp (Crowe et al. 1994a, b, 1995; Whitehead et al. 1998b). Moreover, rRSV-Long/A2cpts and rRSV-Long/A2cptsΔSH share the same ts phenotype and shut-off temperature of around 37 °C, similar to their cpRSV derivative counterparts, cpts-248/404 and rA2cp248/404ΔSH (Crowe et al. 1994a, 1995; Firestone et al. 1996; Karron et al. 2005; Lin et al. 2006; Schickli et al. 2012). We have monitored all the att mutation sites including the set of five cp mutations (Whitehead et al. 1998b) and ts mutations at the 248 and 404 ts markers in the serially passaged rRSVs from P1 to P9, and no reversion was observed at these sites.
However, during the temperature stress test of rRSV-Long/A2cpts, all the randomly selected tsi viruses had nt changes causing reversion to the wild-type amino acid or nt at the 248 or 404 ts markers. According to a previous in vitro analysis of MEDI-559 from MedImmune, an updated version of rA2cp248/404/1030ΔSH and containing 39 silent nt substitutions (including TTA to CTG substitution at the 248 ts marker), MEDI-599 displayed a similar nt change at the 248 site as the resultant tsi viruses (Schickli et al. 2012). In contrast to our observation, no changes were observed at the 404 ts marker located in MEDI-559 and no MEDI-559-infected plates showed positive RSV immunostaining at 38 °C. This phenomenon may be attributed to both the lower shut-off temperature of MEDI-559 (35 °C) and the lower passage temperature (37 °C) used in the cited study than the 39 °C passage temperature applied here for rRSV-Long/A2cpts. Although no changes were reported at the MEDI-559 404 ts, reversion at the 404 locus is possible and did occur during the earlier clinical trials with cpts248/404 in young infants (Whitehead et al. 1998a; Wright et al. 2000). In our analysis of nt changes at the ts markers, cDNA from only five randomly chosen positive wells was sequenced and analyzed for the presence of gene fragments containing ts mutations. Despite its convenience, our method is not as comprehensive and accurate as high-throughput deep sequencing of the whole genomes. Therefore, further analysis might be needed to uncover some of the mutations.
The characteristics of the in vivo att phenotypes of rRSVs were uncovered by infecting mice intranasally. Compared with wtRSV, all the three rRSVs had lower replication activity in vitro (Fig. 2 and Fig. 3), indicating that the cp mutation endowed all the three rRSVs with att phenotype from ‘host-range’ restrictions functioning only in vivo but not in vitro, similar to cpRSV in chimpanzees and humans (Whitehead et al. 1998a, b). Furthermore, the titers of the two rRSVs with ts mutations were reduced in the lower respiratory tract compared with those in the upper respiratory tract (Table 1), in sharp contrast with the titers of rRSV-Long/A2cp. These results demonstrate that restricted replication at physiological temperatures is attributed to the ts phenotypes of rRSV-Long/A2cpts and rRSV-Long/A2cptsΔSH. Moreover, similar levels of serum IgG and neutralizing antibodies were found among mice vaccinated with rRSV-Long/A2cp, rRSV-Long/A2cptsΔSH, and wtRSV (P > 0.05). However, despite the shared ts phenotype, rRSV-Long/A2cpts immunization, unlike rRSV-Long/A2cptsΔSH immunization, resulted in a slightly decreased antibody response compared to wtRSV immunization (P < 0.05). This result slightly differs from that of a previous report (Whitehead et al. 1999), possibly owing to an increased transcription of the downstream neighboring genes G and F in ΔSH rRSV (Bukreyev et al. 1997). Notably, despite the inconsistency in comparison with wtRSV, there were no significant differences in IgG and neutralizing antibody responses among the three rRSVs, similar to the previous observations in chimpanzees (Whitehead et al. 1999).
The most dramatic reduction of lung viral loads occurred in mice vaccinated with rRSV-Long/A2cptsΔSH among the three rRSVs (P < 0.05). Similar results were observed for wtRSV-immunized mice. Therefore, compared with the other two rRSVs, rRSV-Long/A2cptsΔSH exhibited the best protective properties against RSV infection. Finally, analyses of lung pathology and eosinophilia in mice after viral challenge showed the absence of ERD following immunization by either of the rRSVs, especially by rRSV-Long/A2cptsΔSH. These results are likely attributed to the potent efficacy of rRSV-Long/A2cptsΔSH immunization. FI-RSV immunization is known to be followed by ERD; however, there are still uncertainties regarding the underlying mechanism of this phenomenon. Generally, the key factors are thought to be the poor functional activity of the induced RSV-specific antibodies and a Th2-biased CD4 T-cell response characterized by increasing cytokine levels. Previous studies have already confirmed that ERD does not occur after a natural RSV infection or inoculation with RSV LAV candidates (Wright et al. 2007), consistent with our results.
Altogether, rRSVs bearing either cp, cpts, or cptsΔSH mutations have been successfully constructed and rescued from a parent strain other than A2 (wtRSV Long). These data indicate that the proposed strategy allows to develop promising RSV LAV candidates, with att phenotype, immunogenicity, genetic stability, and safety similar to those characteristic for strains derived from the traditional RSV A2 parent. Although it is necessary to confirm these data in animal models more susceptible to human strains of RSV, such as cotton rats, which are 100-fold more permissive than BALB/c mice (Prince et al. 1978, 1999; Byrd and Prince 1997; Taylor 2017), our work has paved the way to improve the safety, immunogenicity, and efficacy of live attenuated RSV vaccine candidates derived from the wtRSV Long parent.
In summary, we have successfully constructed and rescued three rRSVs bearing cp, cpts, or cptsΔSH mutations based on a reverse genetics technology. We used the wtRSV Long parental strain, which differs from the standard RSV A2 strain. However, the three cDNA-derived rRSVs exhibited biological and immunological similarities with their RSV A2 counterparts. Moreover, the results of in vitro and in vivo experiments showed that the rRSVs with the introduced mutations (cp, 248ts, and 404ts) from the RSV A2 strain exhibited optimal levels of attenuation, efficacy, and genetic stability, which could be further improved when combined with the SH gene deletion. Therefore, we can preliminarily conclude that the development of live attenuated RSV vaccines with RSV Long as parent strain is a promising strategy which needs to be further explored in the future.
References
Blanco JC, Boukhvalova MS, Pletneva LM, Shirey KA, Vogel SN (2014) A recombinant anchorless respiratory syncytial virus (RSV) fusion (F) protein/monophosphoryl lipid A (MPL) vaccine protects against RSV-induced replication and lung pathology. Vaccine 32:1495–1500
Bouillier C, Rincheval V, Sitterlin D, Blouquit-Laye S, Desquesnes A, Eleouet JF, Gault E, Rameix-Welti MA (2019) Generation, amplification, and titration of recombinant respiratory syncytial viruses. J Vis Exp. https://doi.org/10.3791/59218
Bukreyev A, Belyakov IM, Berzofsky JA, Murphy BR, Collins PL (2001) Granulocyte-macrophage colony-stimulating factor expressed by recombinant respiratory syncytial virus attenuates viral replication and increases the level of pulmonary antigen-presenting cells. J Virol 75:12128–12140
Bukreyev A, Whitehead SS, Murphy BR, Collins PL (1997) Recombinant respiratory syncytial virus from which the entire SH gene has been deleted grows efficiently in cell culture and exhibits site-specific attenuation in the respiratory tract of the mouse. J Virol 71:8973–8982
Byrd LG, Prince GA (1997) Animal models of respiratory syncytial virus infection. Clin Infect Dis 25:1363–1368
Collins PL, Bermingham A (1999) The M2–2 protein of human respiratory syncytial virus is a regulatory factor involved in the balance between RNA replication and transcription. Proc Natl Acad Sci USA 96:11259–11264
Connors M, Crowe JE, Murphy BR, Firestone CY, Collins PL (1995) A cold-passaged, attenuated strain of human respiratory syncytial virus contains mutations in the F and L genes. Virology 208:478–484
Crowe JE Jr, Bui PT, Davis AR, Chanock RM, Murphy BR (1994a) A further attenuated derivative of a cold-passaged temperature-sensitive mutant of human respiratory syncytial virus retains immunogenicity and protective efficacy against wild-type challenge in seronegative chimpanzees. Vaccine 12:783–790
Crowe JE Jr, Bui PT, London WT, Davis AR, Hung PP, Chanock RM, Murphy BR (1994b) Satisfactorily attenuated and protective mutants derived from a partially attenuated cold-passaged respiratory syncytial virus mutant by introduction of additional attenuating mutations during chemical mutagenesis. Vaccine 12:691–699
Crowe JE Jr, Bui PT, Siber GR, Elkins WR, Chanock RM, Murphy BR (1995) Cold-passaged, temperature-sensitive mutants of human respiratory syncytial virus (RSV) are highly attenuated, immunogenic, and protective in seronegative chimpanzees, even when RSV antibodies are infused shortly before immunization. Vaccine 13:847–855
Crowe JE Jr, Collins PL, London WT, Chanock RM, Murphy BR (1993) A comparison in chimpanzees of the immunogenicity and efficacy of live attenuated respiratory syncytial virus (RSV) temperature-sensitive mutant vaccines and vaccinia virus recombinants that express the surface glycoproteins of RSV. Vaccine 11:1395–1404
Falsey AR, Hennessey PA, Formica MA, Cox C, Walsh EE (2005) Respiratory syncytial virus infection in elderly and high-risk adults. N Engl J Med 352:1749–1759
Firestone CY, Whitehead SS, Collins PL, Murphy BR, Crowe JE Jr (1996) Nucleotide sequence analysis of the respiratory syncytial virus subgroup A cold-passaged (cp) temperature sensitive (ts) cpts-248/404 live attenuated virus vaccine candidate. Virology 225:419–422
Fu YH, He JS, Qiao W, Jiao YY, Hua Y, Zhang Y, Peng XL, Hong T (2013) Intranasal immunization with a helper-dependent adenoviral vector expressing the codon-optimized fusion glycoprotein of human respiratory syncytial virus elicits protective immunity in BALB/c mice. Virol J 10:183
Fu YH, Jiao YY, He JS, Giang GY, Zhang W, Yan YF, Ma Y, Hua Y, Zhang Y, Peng XL, Shi CX, Hong T (2014) Sublingual administration of a helper-dependent adenoviral vector expressing the codon-optimized soluble fusion glycoprotein of human respiratory syncytial virus elicits protective immunity in mice. Antiviral Res 105:72–79
Hu B, Jiang J, Zhan J, Li G, Jiang Y, Guan X, Chen Y, Fang Z (2014) Development of a reverse genetics system for respiratory syncytial virus long strain and an immunogenicity study of the recombinant virus. Virol J 11:142
Huang Y, Cyr SL, Burt DS, Anderson R (2009) Murine host responses to respiratory syncytial virus (RSV) following intranasal administration of a Protollin-adjuvanted, epitope-enhanced recombinant G protein vaccine. J Clin Virol 44:287–291
Jiao YY, Fu YH, Yan YF, Hua Y, Ma Y, Zhang XJ, Song JD, Peng XL, Huang J, Hong T, He JS (2017) A single intranasal administration of virus-like particle vaccine induces an efficient protection for mice against human respiratory syncytial virus. Antiviral Res 144:57–69
Johnson PR, Spriggs MK, Olmsted RA, Collins PL (1987) The G glycoprotein of human respiratory syncytial viruses of subgroups A and B: extensive sequence divergence between antigenically related proteins. Proc Natl Acad Sci USA 84:5625–5629
Jorquera PA, Anderson L, Tripp RA (2016) Understanding respiratory syncytial virus (RSV) vaccine development and aspects of disease pathogenesis. Expert Rev Vaccines 15:173–187
Kapikian AZ, Mitchell RH, Chanock RM, Shvedoff RA, Stewart CE (1969) An epidemiologic study of altered clinical reactivity to respiratory syncytial (RS) virus infection in children previously vaccinated with an inactivated RS virus vaccine. Am J Epidemiol 89:405–421
Karron RA, Wright PF, Belshe RB, Thumar B, Casey R, Newman F, Polack FP, Randolph VB, Deatly A, Hackell J, Gruber W, Murphy BR, Collins PL (2005) Identification of a recombinant live attenuated respiratory syncytial virus vaccine candidate that is highly attenuated in infants. J Infect Dis 191:1093–1104
Kim HW, Canchola JG, Brandt CD, Pyles G, Chanock RM, Jensen K, Parrott RH (1969) Respiratory syncytial virus disease in infants despite prior administration of antigenic inactivated vaccine. Am J Epidemiol 89:422–434
Kohlmann R, Schwannecke S, Tippler B, Ternette N, Temchura VV, Tenbusch M, Uberla K, Grunwald T (2009) Protective efficacy and immunogenicity of an adenoviral vector vaccine encoding the codon-optimized F protein of respiratory syncytial virus. J Virol 83:12601–12610
Kwanten LC, De Clerck B, Roymans D (2013) A fluorescence-based high-throughput antiviral compound screening assay against respiratory syncytial virus. In: Gong EY (ed) Antiviral methods and protocols, methods in molecular biology, vol 1030. Springer Science, Berlin, pp 337–344
Lawlor HA, Schickli JH, Tang RS (2013) A single amino acid in the F2 subunit of respiratory syncytial virus fusion protein alters growth and fusogenicity. J Gen Virol 94:2627–2635
Lin YH, Deatly AM, Chen W, Miller LZ, Lerch R, Sidhu MS, Udem SA, Randolph VB (2006) Genetic stability determinants of temperature sensitive, live attenuated respiratory syncytial virus vaccine candidates. Virus Res 115:9–15
Luongo C, Winter CC, Collins PL, Buchholz UJ (2013) Respiratory syncytial virus modified by deletions of the NS2 gene and amino acid S1313 of the L polymerase protein is a temperature-sensitive, live-attenuated vaccine candidate that is phenotypically stable at physiological temperature. J Virol 87:1985–1996
Ma Y, Jiao YY, Yu YZ, Jiang N, Hua Y, Zhang XJ, Fu YH, Peng XL, Zheng YP, Anderson LJ, He JS (2018) A built-in CpG adjuvant in RSV F protein DNA vaccine drives a Th1 polarized and enhanced protective immune response. Viruses 10:38
Magro M, Mas V, Chappell K, Vazquez M, Cano O, Luque D, Terron MC, Melero JA, Palomo C (2012) Neutralizing antibodies against the preactive form of respiratory syncytial virus fusion protein offer unique possibilities for clinical intervention. Proc Natl Acad Sci USA 109:3089–3094
McFarland EJ, Karron RA, Muresan P, Cunningham CK, Libous J, Perlowski C, Thumar B, Gnanashanmugam D, Moye J, Schappell E, Barr E, Rexroad V, Fearn L, Spector SA, Aziz M, Cielo M, Beneri C, Wiznia A, Luongo C, Collins P, Buchholz UJ (2020a) Live respiratory syncytial virus attenuated by m2–2 deletion and stabilized temperature sensitivity mutation 1030s is a promising vaccine candidate in children. J Infect Dis 221:534–543
McFarland EJ, Karron RA, Muresan P, Cunningham CK, Perlowski C, Libous J, Oliva J, Jean-Philippe P, Moye J, Schappell E, Barr E, Rexroad V, Fearn L, Cielo M, Wiznia A, Deville JG, Yang L, Luongo C, Collins PL, Buchholz UJ (2020b) Live-attenuated respiratory syncytial virus vaccine with m2–2 deletion and with small hydrophobic noncoding region is highly immunogenic in children. J Infect Dis 221:2050–2059
McFarland EJ, Karron RA, Muresan P, Cunningham CK, Valentine ME, Perlowski C, Thumar B, Gnanashanmugam D, Siberry GK, Schappell E, Barr E, Rexroad V, Yogev R, Spector SA, Aziz M, Patel N, Cielo M, Luongo C, Collins PL, Buchholz UJ, International Maternal Pediatric Adolescent ACTST (2018) Live-attenuated respiratory syncytial virus vaccine candidate with deletion of RNA synthesis regulatory protein M2–2 is highly immunogenic in children. J Infect Dis 217:1347–1355
PERCH G (2019) Causes of severe pneumonia requiring hospital admission in children without HIV infection from Africa and Asia: the PERCH multi-country case-control study. Lancet 394:757–779
Peret TC, Hall CB, Schnabel KC, Golub JA, Anderson LJ (1998) Circulation patterns of genetically distinct group A and B strains of human respiratory syncytial virus in a community. J Gen Virol 79:2221–2229
Prince GA, Jenson AB, Horswood RL, Camargo E, Chanock RM (1978) The pathogenesis of respiratory syncytial virus infection in cotton rats. Am J Pathol 93:771–791
Prince GA, Prieels JP, Slaoui M, Porter DD (1999) Pulmonary lesions in primary respiratory syncytial virus infection, reinfection, and vaccine-enhanced disease in the cotton rat (Sigmodon hispidus). Lab Invest 79:1385–1392
Rostad CA, Stobart CC, Gilbert BE, Pickles RJ, Hotard AL, Meng J, Blanco JCG, Moin SM, Graham BS, Piedra PA, Moore ML (2016) A recombinant respiratory syncytial virus vaccine candidate attenuated by a low-fusion F protein is immunogenic and protective against challenge in cotton rats. J Virol 90:7508–7518
Schickli JH, Kaur J, Tang RS (2012) Nonclinical phenotypic and genotypic analyses of a Phase 1 pediatric respiratory syncytial virus vaccine candidate MEDI-559 (rA2cp248/404/1030DeltaSH) at permissive and non-permissive temperatures. Virus Res 169:38–47
Singh SR, Dennis VA, Carter CL, Pillai SR, Jefferson A, Sahi SV, Moore EG (2007) Immunogenicity and efficacy of recombinant RSV-F vaccine in a mouse model. Vaccine 25:6211–6223
Taylor G (2017) Animal models of respiratory syncytial virus infection. Vaccine 35:469–480
Teng MN, Mejias A, Ramilo O, Peeples ME (2020) Live attenuated vaccine with a stabilized mutation and gene deletion for prevention of respiratory syncytial virus disease in young children. J Infect Dis 221:501–503
Troeger C, Blacker B, Khalil IA, Rao PC, Cao J, Zimsen SRM, Albertson SB, Deshpande A, Farag T, Abebe Z et al. (2018) Estimates of the global, regional, and national morbidity, mortality, and aetiologies of lower respiratory infections in 195 countries, 1990–2016: a systematic analysis for the Global Burden of Disease Study 2016. Lancet Infect Dis 18:1191–1210
van Elden LJ, van Loon AM, van der Beek A, Hendriksen KA, Hoepelman AI, van Kraaij MG, Schipper P, Nijhuis M (2003) Applicability of a real-time quantitative PCR assay for diagnosis of respiratory syncytial virus infection in immunocompromised adults. J Clin Microbiol 41:4378–4381
Whitehead SS, Bukreyev A, Teng MN, Firestone CY, St Claire M, Elkins WR, Collins PL, Murphy BR (1999) Recombinant respiratory syncytial virus bearing a deletion of either the NS2 or SH gene is attenuated in chimpanzees. J Virol 73:3438–3442
Whitehead SS, Firestone CY, Collins PL, Murphy BR (1998a) A single nucleotide substitution in the transcription start signal of the M2 gene of respiratory syncytial virus vaccine candidate cpts248/404 is the major determinant of the temperature-sensitive and attenuation phenotypes. Virology 247:232–239
Whitehead SS, Juhasz K, Firestone CY, Collins PL, Murphy BR (1998b) Recombinant respiratory syncytial virus (RSV) bearing a set of mutations from cold-passaged RSV is attenuated in chimpanzees. J Virol 72:4467–4471
Wright PF, Karron RA, Belshe RB, Thompson J, Crowe Jr JE, Boyce TG, Halburnt LL, Reed GW, Whitehead SS, Anderson EL, Wittek AE, Casey R, Eichelberger M, Thumar B, Randolph VB, Udem SA, Chanock RM, Murphy BR (2000) Evaluation of a live, cold-passaged, temperature-sensitive, respiratory syncytial virus vaccine candidate in infancy. J Infect Dis 182:1331–1342
Wright PF, Karron RA, Belshe RB, Shi JR, Randolph VB, Collins PL, O’Shea AF, Gruber WC, Murphy BR (2007) The absence of enhanced disease with wild type respiratory syncytial virus infection occurring after receipt of live, attenuated, respiratory syncytial virus vaccines. Vaccine 25:7372–7378
Xu M, Jiao YY, Fu YH, Jiang N, Zheng YB, Yan YF, Zhang M, Zheng YP, Zhu WY, Peng XL, He JS (2018) Construction and characterization of a recombinant human respiratory syncytial virus encoding enhanced green fluorescence protein for antiviral drug screening assay. Biomed Res Int 2018:8431243
Acknowledgements
We thank Dr. Prof. Jean-François Eléouët, from Virologie et Immunologie Moléculaires (VIM), French National Institute for Agriculture, Food, and Environment (INRAE), Paris, France and Marie-Anne Rameix-Welti, from UFR des Sciences de la Santé Simone Veil, Université de Versailles Saint-Quentin, Versailles, France for kind suggestion and support. This work was supported by grants from the Natural Science Foundation of China (81771777, 32070922).
Author information
Authors and Affiliations
Contributions
JSH conceived and designed the study. LNW, MX, YBZ, YYJ, and XLP conducted the experiments. LNW, MX, JMY, YHF, YPZ, WYZ, and ZJD analyzed the data. XLP and LNW drafted and revised the paper. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflicts of interest.
Human and Animal Rights
This study was conducted with the approval of the Institutional Animal Care and Use Committees of Tsinghua University (No. 17-DZJ1).
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Wang, LN., Peng, XL., Xu, M. et al. Evaluation of the Safety and Immune Efficacy of Recombinant Human Respiratory Syncytial Virus Strain Long Live Attenuated Vaccine Candidates. Virol. Sin. 36, 706–720 (2021). https://doi.org/10.1007/s12250-021-00345-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12250-021-00345-3