Abstract
Co-occurring predators often exhibit ecological niche partitioning, resulting from competition over evolutionary time. However, in productive estuarine ecosystems with high resource availability, predators may occupy similar niches without conflict. Determining the degree of niche partitioning and overlap among co-occurring predators can provide insights into a food web’s function and its potential resiliency to perturbations. This study used stable isotope analysis to assess the trophic ecology of four predators in Galveston Bay, Texas, USA: spotted seatrout, black drum, bull shark, and alligator gar. Spatially distinct primary producer isotopic ratios emerged for both δ13C and δ15N following salinity regimes, which translated to similar patterns in predator tissue. The volume and overlap among species’ trophic niches also varied spatially, with species-specific expansion and contraction of niches across the freshwater-marine continuum. The observed niche patterns were likely related to movements, with implications for trophic coupling across the estuarine landscape. Using regional delineations for baseline values yielded trophic position estimates that were validated by compound-specific stable isotopes and were similar (3.77 to 3.96) for all species but black drum (3.25). Trophic position increased with body length for all species but black drum, and these relationships differed when using estuary-wide versus regionally distinct baselines. Alligator gar gut contents were examined, which primarily aligned with piscivory but also included previously unreported taxa (insect, mammal). Collectively, these results provide evidence for spatial and ontogenetic shifts in trophic ecology within this predator assemblage and highlight the importance of spatial scale when using stable isotopes to examine estuarine food webs.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Estuaries are among the most productive ecosystems on the planet, with nutrient inputs from natural and anthropogenic sources fueling primary production that forms the base of complex food webs (Cloern et al. 2014). Many predator species co-occur in estuaries, sharing habitats and food resources, and some degree of specialization in trophic ecology is expected to emerge over evolutionary timescales to reduce competitive pressures among species with similar habitat requirements (MacArthur 1958; Hutchinson 1959). The resulting resource partitioning allows predators to coexist spatiotemporally while they occupy slightly different realized niches (Hartman and Brandt 1995; Matich et al. 2011; Kroetz et al. 2016). However, since prey resources are often abundant in these ecosystems, minimal direct competition among predators can allow for similarities in their trophic ecology (Walker 1992). This redundancy within trophic guilds, such as among top predators, can in turn stabilize the ecosystem as a whole in the face of perturbations (Naeem 1998; Sanders et al. 2018; Biggs et al. 2020). While perfect redundancy and complete trophic partitioning are unlikely (Loreau 2004), exploring the interplay between these concepts can aid in understanding both food web dynamics and the potential resiliency of a system to environmental and anthropogenic stressors.
Trophic ecology, and therefore niche partitioning and overlap, is inherently linked to the movement patterns of predators both within and among species. Large-bodied predators situated at high trophic levels tend to have greater scales of movement, and this mobility allows them to respond to changes in the spatiotemporal arrangement of resources (Rooney et al. 2008). Foraging movements can therefore connect otherwise disparate habitats across landscapes, which act to stabilize food webs by dampening spatiotemporal fluctuations in prey populations and provide spatial subsidies of energy and nutrients (Polis et al. 1997; McCann et al. 2005; Rooney et al. 2006). Furthermore, conspecific individuals can exhibit dramatically different trophic niches due to the wide array of intrinsic and extrinsic factors they each experience, including morphological and behavioral traits and environmental characteristics (Bolnick et al. 2003). Both inter- and intraspecific diversities in trophic ecology influence the stability and function of ecosystems (Schmitz 2009; Lefcheck and Duffy 2015; Allgeier et al. 2020), so effective conservation and management likely requires employing strategies that maintain or enhance predator functional diversity (Clemente et al. 2010; Cardinale et al. 2012).
Stable isotope analysis (SIA) is a powerful tool commonly used to assess aspects of the trophic ecology of predators (Peterson and Fry 1987; Layman et al. 2012). Nitrogen stable isotopes (ratio of 15N: 14N, δ15N) undergo significant fractionation, becoming enriched at a fairly consistent rate with each trophic transfer, and can therefore be used to determine the trophic level of a predator (Minagawa and Wada 1984; Post 2002). Fractionation is minimal for carbon stable isotopes (ratio of 13C: 12C, δ13C) compared to nitrogen, so δ13C is used to infer the basal sources of a predator’s food web (DeNiro and Epstein 1978). Current analytical methods for stable isotope ecology often incorporate Bayesian stable isotope mixing models, which estimate the proportional contributions of various basal resources or prey to a consumer’s diet (Stock and Semmens 2016; Parnell et al. 2010). These models are crucial for deciphering complex feeding relationships within ecological communities (Ward et al. 2010; Newsome et al. 2012; Stock et al. 2018). Recently, hypervolume analysis based on dietary contribution estimates from mixing models has been employed by researchers to assess niche volume and overlap among predators (Rezek et al. 2020; James et al. 2020). Hypervolume analysis, a concept grounded in stochastic geometry, quantifies the multidimensional ecological niche of an organism (Blonder 2014). This approach provides a comprehensive understanding of trophic niche dynamics through high-dimensional kernel density estimation of isotope-based ecological data. However, there are many factors that can influence isotopic ratios of consumers such as diet quality, nutritional condition, tissue turnover rates, and habitat use (Hette-Tronquart 2019; Shipley and Matich 2020). It is therefore important to evaluate isotopic ratios across space, time, and ontogeny and to draw upon previous literature describing the habitat use patterns and diets of consumers to interpret trophic position estimates and trophic niche size and overlap.
SIA relies on knowledge of baseline isotopic values of primary producers or consumers to accurately estimate trophic position (TP), which is often difficult to resolve as they vary spatiotemporally (Grey et al. 2001; McMahon et al. 2013). To alleviate this issue, compound-specific isotope analysis of amino acids (CSIA-AA) has been increasing in popularity as a powerful addition to SIA. CSIA-AA does not require sampling of baseline isotopic values to delineate the trophic structure of an ecosystem. Instead, δ15N values from specific amino acids are isolated from predator tissue that are either considered “source” or “trophic” based on known fractionation behaviors (Popp et al. 2007). Source amino acids (e.g., phenylalanine) are those that exhibit very little fractionation during trophic transfer and have been found to accurately represent the isotopic ratios of organisms at the base of the food web over space and time (McClelland and Montoya 2002). Trophic amino acids (e.g., glutamic acid) undergo fractionation due to transamination and deamination, resulting in enriched δ15N values relative to the source amino acids (Popp et al. 2007; Chikaraishi et al. 2009). By comparing the isotopic values of source and trophic amino acids within an individual sample, TP of the predator can be determined without additional sampling for baseline values (Hetherington et al. 2018). Assessing TP using either bulk SIA or CSIA-AA requires estimating the degree of N enrichment that occurs with each successive increase in trophic level (trophic enrichment factor), which can be affected by prey quality, N metabolism/excretion pathways, and other intrinsic and extrinsic variables (Nielsen et al. 2015). These complexities can introduce uncertainty to trophic position estimation, but the accuracy of CSIA-AA-derived TP estimates increases when multiple source and trophic amino acids are incorporated (Bradley et al. 2015).
Use of SIA in estuarine ecosystems is also often complicated by dramatic spatial variation in stable isotope ratios of both plants and animals along the freshwater-marine continuum (Peterson and Fry 1987). Sources of inorganic carbon and dominant photosynthetic pathways (C3 vs. C4) differ between terrestrial, freshwater, and marine systems, which generally results in depleted baseline δ13C values in the low-salinity regions of an estuary. This connection between the estuary and its freshwater source(s) also influences δ15N ratios, as excess ammonium and nitrate enter the system via anthropogenic inputs (e.g., runoff and wastewater) and enriches the ambient δ15N baseline (McClelland et al. 1997; Bishop et al. 2017). These spatial patterns of stable isotope ratios can translate to the isotopic ratios of higher-order consumers (Matich et al. 2021), which, if left unaccounted for, can lead to erroneous conclusions regarding the consumer’s trophic ecology.
In addition to spatial differences in stable isotope ratios, many predator species exhibit ontogenetic (temporal) changes in their diets and habitat use patterns. These shifts can result in increasing trophic position as the animal grows larger, due to a variety of factors such as increased gape size and the ability to forage across larger spatial scales (Galván et al. 2010; Grubbs et al. 2010). The trophic position of an organism is an integral metric for understanding the role they play in their ecosystem, so it is important to examine whether that role changes over their lifespan. Furthermore, trophic position estimates using bulk SIA are sensitive to baseline δ15N values (Woodcock et al. 2012), which are especially important in heterogeneous ecosystems like estuaries. Comparing these estimates with CSIA-AA-derived estimates can provide validation, since environmentally sampled baselines are not required.
This study examined the trophic ecology of four euryhaline predators in the Galveston Bay Complex (GBC), Texas, USA: spotted seatrout (Cynoscion nebulosus), black drum (Pogonias cromis), bull shark (Carcharhinus leucas), and alligator gar (Atractosteus spatula). These species all inhabit estuaries in the northern Gulf of Mexico (GOM) but exemplify the diversity of predators in these ecosystems, differing notably in their habitat use patterns and salinity preferences: spotted seatrout are the least freshwater-associated, bull sharks and black drum are highly euryhaline, and alligator gar are the most freshwater-associated (Livernois et al. 2021). Furthermore, most of these predators are considered primarily piscivorous (Simonsen and Cowan 2013; TinHan and Wells 2021; Marsaly et al. 2023), with the exception of black drum which feed on benthic invertebrates (Rubio et al. 2018). Using these four species as representative estuarine predators, the objectives of this study are to employ bulk SIA and CSIA-AA to (1) examine spatial patterns of baseline and predator isotopic ratios in the GBC, (2) describe the trophic niche area and degree of overlap among species using Bayesian mixing model-derived source contribution estimates, (3) estimate the trophic position of each species using SIA and CSIA-AA, and (4) examine potential ontogenetic shifts in trophic position.
Methods
Study Area and Hydrology
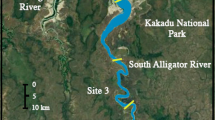
The GBC is a large (1420 km2 surface area) estuary located in the subtropical northwestern Gulf of Mexico. The system hosts diverse fish assemblages (asymptotic Shannon index ~ 20; Pawluk et al. 2021) and primary producers including seagrass, saltmarsh (primarily Spartina alterniflora), phytoplankton, and freshwater-associated macrophytes. Distinct spatial patterns exist for salinity in the estuary, based primarily on the hydrology of riverine and marine inputs. The main sources of freshwater input to the GBC are the Trinity River (55%) and San Jacinto River (26%), which enter in the northern reaches of the estuary (Trinity Bay, Fig. 1; Guthrie et al. 2012). Trinity Bay receives the majority of this freshwater input, a portion of which subsequently flows into East Bay where it mixes with marine water entering through the Galveston Ship Channel (Powell et al. 2003). West Bay is hydrologically separated from the majority of freshwater input by the Texas City Dike and thus receives most exchange through passes to the Gulf of Mexico (Powell et al. 2003; Fig. 1). Freshwater enters West Bay through Chocolate Bayou, but this inflow is relatively small, only contributing approximately 4% of freshwater input to the GBC (Guthrie et al. 2012).
Fish Sample Collection
Individuals of each predator species were collected during fall 2020 and spring 2021 by the Texas Parks and Wildlife Department (TPWD) Coastal Fisheries Division. Sampling occurred across two seasons to assess potential biases related to temporal changes in ambient isotopic values. During TPWD’s long-term Marine Resource Monitoring Program sampling efforts (Martinez-Andrade et al. 2009), fishes were collected from gillnets and subsequently frozen whole at – 20 °C. Forty-five gillnets were set each season, with locations being determined by a stratified cluster sampling method that ensures coverage of the full extent of the estuary. Collection location (latitude/longitude) and a suite of physicochemical conditions were recorded during each gillnet set. Thirty individuals of each species were collected, with approximately half collected in fall 2020 and the other half in spring 2021. Within each season, individuals were collected as evenly as possible across the GBC (Fig. 2a–d and Table 1). Prior to stable isotope analysis, frozen fishes were thawed, and a biopsy of white muscle tissue (2–3 cm3) was extracted from the left dorsal region. Muscle samples were subsequently frozen at – 20 °C until processing.
To prepare each muscle sample for SIA, a small section (~ 1 cm3) was dissected to remove any skin, connective tissue, and bone and was thoroughly rinsed with deionized (DI) water. Cleaned muscle samples were then oven-dried at 60 °C for 48–72 h and homogenized with a mortar and pestle. Lipid and urea were extracted from dried muscle samples to account for potential differences in their concentrations among species, which can confound the interpretation of carbon stable isotope ratios (Post et al. 2007). Following Kim and Koch (2012), lipids and urea were extracted from dried, homogenized muscle tissue using petroleum ether and DI water, respectively, with a Dionex Automated Solvent Extractor (ASE, Dionex). Each sample was packed in an ASE cell between pre-combusted (450 °C for 4 h) 30 mm GF/B filters, and any remaining space was filled with clean sand. The cell was then rinsed three times with 100% petroleum ether at 100 °C, 1500 psi, and 60% rinse volume for 5 min, followed by three rinses with DI water using the same settings (Plumlee et al. 2021). Samples were removed from the cell, oven-dried at 60 °C for 12–24 h, and homogenized with a mortar and pestle. Finally, ~ 1.2 mg of each sample was packaged in 5 × 9 mm tin capsules and shipped to the University of California–Davis Stable Isotope Facility (UC-Davis SIF) for analysis.
Primary Producer Sample Collection
To estimate the isotopic baseline of the ecosystem, samples of particulate organic matter (POM), saltmarsh grass (Spartina alterniflora), seagrasses (Halodule wrightii, Thalassia testudinum), and freshwater macrophytes (Phragmites australis, Typha spp., Carex spp.) were collected once per fall and spring season to correspond with predator collections. These primary producers were collected from 11 sites (where present) distributed throughout the GBC to assess regional differences in baseline isotopic ratios (Fig. 2e). Location and physicochemical conditions (temperature, salinity, dissolved oxygen) were recorded at each collection site. For POM, 1 L of water was collected from the surface at all sites once per season (n = 22). Water samples were pre-filtered across a 63-µm Nitex mesh screen to remove zooplankton and large debris, followed by filtering across pre-combusted 47 mm GF/F filters. Each filter was then oven-dried at 60 °C for 24 h, cut in half, weighed, and packaged in tin capsules. Macrophytes (Spartina: n = 14 from 7 sites, seagrass: n = 5 from 2 sites, and freshwater plants: n = 8 from 2 sites) were frozen whole at – 20 °C before processing. Aboveground biomass (live, undamaged material) was isolated from one plant per sample, which was subsequently rinsed and agitated with DI water to remove debris and epiphytes. Plant material was oven-dried at 50 °C for up to 1 week, which allowed the material to become brittle and more easily homogenized. Dried plant material was homogenized using an electric coffee grinder and/or a mortar and pestle, and ~ 3.7 mg of each sample was packaged in 5 × 9-mm tin capsules. All primary producer samples were shipped to the UC-Davis SIF for analysis with fish muscle samples.
Bulk Stable Isotope Analysis
At the UC-Davis SIF, the abundance of 12C, 13C, 14N, and 15N in each sample (fish muscle and primary producers) was determined using an Elementar vario MICRO cube elemental analyzer interfaced to an Elementar VisION isotope ratio mass spectrometer (Elementar Analysensysteme GmbH, Langenselbold, Germany). Ratios of 13C:12C and 15N:14N are reported in delta notation using the following equation:
where Rsample is the ratio of the heavy to light isotope in the sample and Rstandard is the ratio of the heavy to light isotope derived from the accepted international standards, Vienna Pee Dee Belemnite for carbon and atmospheric N2 for nitrogen. The average standard deviation of reference materials for δ13C and δ15N was 0.04‰ and 0.09‰, respectively.
Compound-Specific Stable Isotope Analysis of Amino Acids
Given the high cost of analysis and increased accuracy of CSIA-AA (Chikaraishi et al. 2009), a small subset (n = 6 per species) of the bulk SIA samples was used, with individuals of each species distributed as evenly as possible between seasons and regions (Table 1). Sample preparation for CSIA-AA followed the same procedure as described for bulk SIA, but lipid extraction was not conducted. Dried, homogenized muscle (~ 2.8 mg) was packaged in clean 2-mL glass vials and shipped to the UC-Davis SIF for analysis. The analysis procedure at UC-Davis SIF followed Yarnes and Herszage (2017), whereby amino acids were isolated from sample material by acid hydrolysis and derivatized as N-acetyl methyl esters. These amino acid derivatives were separated on an Agilent DB-35 column by injection at 260 °C (splitless, 1 min) at a constant flow rate of 2 mL/min. Nitrogen stable isotopes (reported in delta notation) of each amino acid derivative were determined using a Thermo Trace GC 1310 gas chromatograph coupled to a Thermo Scientific Delta V Advantage isotope ratio mass spectrometer via a GC IsoLink II combustion interface. Each sample was analyzed in duplicate, to ensure that values did not exceed the expected measurement error (± 1‰). Norleucine and two amino acid compounds developed by UC-Davis SIF were used as internal standards during analysis. The average standard deviation of δ15N of sample amino acids and reference materials was 0.34‰ and 0.53‰, respectively. Weighted mean values of “source” (δ15Nsource) and “trophic” (δ15Ntrophic) amino acids were calculated following Bradley et al. (2015), with weighting based on the standard deviation of each amino acid. Therefore, δ15Nsource represents the weighted mean of the amino acids glycine, lysine, and phenylalanine, while δ15Ntrophic represents the weighted mean of the amino acids alanine, leucine, and glutamic acid (Bradley et al. 2015; Gloeckler et al. 2018). Mean and standard deviation of individual amino acids used to calculate weighted mean δ15Ntrophic and δ15Nsource are listed in Table 2.
Stomach Contents (Alligator Gar)
Prey composition of stomach contents was assessed for alligator gar, considering the limited empirical evidence regarding their diets in estuarine systems compared to the other species. Individuals used for SIA were examined, in addition to other individuals incidentally captured by the same gillnet survey (n = 57). The contents of full stomachs (containing any potentially identifiable material) were preserved in 10% formalin for 48 h and subsequently stored in 70% ethyl alcohol. All contents were visually identified to the lowest taxonomic level possible, and dietary composition was enumerated as % frequency of occurrence (%FO): the number of stomachs containing at least one individual of a given taxon divided by the total number of full stomachs.
Statistical Analysis
Carbon and nitrogen stable isotopes in estuaries similar to the GBC are often influenced by freshwater inflow, with notably enriched baseline δ15N values observed in low-salinity regions (Bishop et al. 2017; Matich et al. 2021). Values of δ13C and δ15N for each primary producer sample were therefore plotted against salinity (Fig. S1), and the GBC was partitioned into three regions that best represented the observed salinity and isotopic baseline regimes (Fig. 1). Two-way analysis of variance (ANOVA) tests were used to quantify differences in salinity, baseline δ13C, and baseline δ15N values among regions and seasons (and the interaction between them), followed by Tukey’s post hoc tests with Westfall p-value adjustment of pairwise differences for statistically significant effects (using packages “car” and “multcomp”; Hothorn et al. 2008; Fox and Weisberg 2019). To determine whether δ13C and δ15N of fish muscle tissue followed a similar pattern among regions as the baseline values, two-way ANOVA tests comparing each isotope among regions and seasons were conducted for each species independently, followed by Tukey’s post hoc tests with Westfall p-value adjustment.
Bayesian stable isotope mixing models were employed to determine the contribution of four groups of primary producers representing unique basal resources throughout the GBC (FW POM & Plants, Marine POM, Marine Plants, FW Spartina; Fig. S2) using the package MixSIAR (v 3.1.12; Semmens et al. 2013). Individual primary producer samples were assigned to basal resource groups based on salinity and sample type: FW POM & Plants includes POM and freshwater macrophytes from 0.1 to 20.4 psu (mean = 7.75 psu), Marine POM includes POM from 14.9 to 29 psu (23.75 psu), Marine Plants includes Spartina and seagrass from 14.9 to 28.1 psu (23.01), and FW Spartina includes Spartina from 0.3 to 20.4 psu (9.35). Mixing models (without concentration dependence) were conducted for each species using individual fish ID as a factor and were run in three chains with 1,000,000 iterations, a burn-in of 500,000, and a thin of 500 to ensure convergence. Trophic fractionation factors of 1.3 ± 0.3 and 3.3 ± 0.26 were used for C and N, respectively, which were multiplied by each species’ mean CSIA-AA-derived trophic position minus one (McCutchan Jr. et al. 2003). Mixing models estimated proportional contributions of each basal resource group to each individual, which were represented as posterior distributions in a series of numerically calculated vectors that incorporate the error present from isotopic measurement and environmental/ecological variability (Newsome et al. 2012). A mean proportional contribution of each basal resource group to each individual fish was calculated from the corresponding posterior distribution, which were z-transformed before further analysis to create standardized, comparable axes in n-dimensional space (Blonder et al. 2014). Relative source contributions were also used to calculate a trophic position (TP) for each individual fish using the following equation:
where ∆δ15N is 3.3‰ (McCutchan et al. 2003), δ15Nind is the N value of an individual consumer, δ15Nsource is the N value of each basal resource group, and mean %contsource is the mean proportional contribution of each basal resource group to the consumer’s diet (Nelson et al. 2015). This calculates a trophic position for each individual of each species relative to the basal resource groups, but not an absolute trophic position.
To determine the trophic niche size and overlap among species in each region of the GBC (Trinity Bay, East Bay, West Bay), the above-calculated z-transformed mean proportional contributions of each basal resource group and relative TP were used to create hypervolumes representing each species’ regional multidimensional trophic niche as a function of dietary contributions (Blonder et al. 2014; Wilson et al. 2017; James et al. 2020). Specifically, the hypervolume package (v 3.1.3; Blonder et al. 2023) was used to seed Gaussian kernel density estimations, generating a cloud of points along the five axes for each species in each region. Each hypervolume included 95% of the total probability density (quantile threshold = 0.05; Blonder et al. 2014). The size of each hypervolume was calculated, representing the relative breadth of each species’ regional trophic niche. The degree of overlap between each pair of species’ hypervolumes (trophic niches) was determined in each region using the Sørensen overlap index (proportion overlapping) and the fraction of unique hypervolume space per species in each pair.
Absolute trophic position (TP) was estimated for each species using bulk SIA and CSIA-AA. For bulk SIA, the following equation was used (Post 2002):
where λ is the trophic level of the baseline (1 for primary producers), δ15Nconsumer is the isotopic value of the predator, δ15Nbase is the isotopic value of the baseline, and TDF is the trophic discrimination factor. TDF was set at 3.4‰ which is a widely applicable and commonly used average value across ecosystems (Post 2002). An overall (pooled) estimate of TP was calculated for each individual of each species using a GBC-wide baseline value, the average δ15N of all primary producers across all regions (7.87‰). However, to account for the observed differences in baseline and fish muscle δ15N across regions, another TP was calculated for each individual of each species using only data from the region in which it was captured. The average δ15N values of primary producers collected in each region were used as that region’s baseline: 9.97‰ in Trinity Bay, 8.45‰ in East Bay, and 5.18‰ in West Bay.
Trophic position was also calculated using CSIA-AA to compare to bulk SIA TP estimates using the following equation:
where δ15Ntrophic represents the weighted mean trophic amino acid value, δ15Nsource represents the weighted mean source amino acid value, β represents the difference in δ15N between trophic and source amino acids of primary producers, and TDFAA is the isotopic enrichment between trophic and source amino acids of consumers. β and TDFAA were set to 3.6‰ and 5.7‰, respectively (Bradley et al. 2015). A two-way ANOVA was conducted to determine differences in TP between calculation method (bulk SIA vs. CSIA-AA), between species, and the interaction between those variables. If no interaction was present, the main effects of each variable were tested, and if significant, followed by Tukey’s post hoc tests with Westfall p-value adjustment.
Ontogenetic shifts in trophic position were examined using linear regression of TP against total length for each species. Two sets of linear regression were performed, one using pooled TP estimates and the other using regional TP estimates. To determine whether there were differences in total length among regions that may confound these results, a one-way ANOVA per species was conducted comparing total length among regions, followed by Tukey’s post hoc tests with Westfall p-value adjustment. All linear models conducted in this study (ANOVA or linear regression) were checked for the assumptions of homoscedasticity and normality of residuals using Levene’s test and Shapiro–Wilk test, respectively. All statistical analyses were performed in R version 4.2.0 (R Core Team 2022).
Results
Bulk SIA Ratios
Across the GBC, primary producer δ13C values ranged from − 31.53 to − 12.86‰ and δ15N values ranged from 0.86 to 13.41‰. Consumer muscle tissue δ13C ranged from − 22.15 to − 18.25‰ for spotted seatrout, − 24.57 to − 16.62‰ for black drum, − 22.17 to − 15.95‰ for bull sharks, and − 22.32 to − 16.20‰ for alligator gar. Consumer δ15N ranged from 14.90 to 21.82‰ for spotted seatrout, 11.21 to 18.59‰ for black drum, 14.86 to 19.56‰ for bull sharks, and 10.56 to 21.56‰ for alligator gar.
Spatiotemporal Assessment of Bulk SIA
Salinity differed among regions (F2,52 = 17.85, p < 0.001; Fig. 3a), with Trinity Bay exhibiting the lowest average salinity (5.91 psu, Tukey contrasts, p < 0.001), followed by East Bay (15.41 psu) and West Bay (20.42 psu, p = 0.061). No interaction was observed between region and season for baseline δ13C or δ15N (δ13C: F2,49 = 0.015, p = 0.99; δ15N: F2,49 = 1.01, p = 0.37), nor did either isotope differ between seasons (δ13C: F1,51 = 0.11, p = 0.74; δ15N: F1,51 = 0.014, p = 0.91; Fig. S3). Baseline δ13C (all primary producers pooled) differed among regions (F2,52 = 12.62, p < 0.001; Fig. 3b): Trinity Bay (− 26.48‰) exhibited a lower average baseline δ13C than both East Bay (− 19.97‰) and West Bay (− 13.36‰, both Tukey contrasts, p < 0.001), but the latter two had overlapping values ranging from − 31.53 to − 12.86 (p = 0.38). Regional differences in baseline δ15N (all primary producers pooled) were also observed (F2,52 = 21.94, p < 0.001; Fig. 3c), with Trinity Bay exhibiting the highest mean δ15N baseline value (9.97‰, Tukey contrasts, p = 0.053 with East Bay and p < 0.001 with West Bay), followed by East Bay (8.45‰) and lastly West Bay (5.18‰, p < 0.001).
Boxplots depicting differences in surface salinity (a), δ13C (b), and δ15N (c) for all primary producer samples (all types grouped) among regions. Boxes represent the 25th percentile, median, and 75th percentile, with whiskers representing 1.5 times the interquartile range. Outliers (data points that fall outside the range of the whiskers) are represented by points. Letters denote statistical differences between regions for each response variable based on ANOVA with Tukey post hoc tests
Similar patterns of δ13C and δ15N emerged among regions for each fish species, implying that differences in baseline values translated to different regional consumer isotope ratios (Fig. 4). As with the baseline, no interaction between region and season was observed for δ13C or δ15N for any fish species, nor were there notable differences between seasons (Fig. S3, details in Tables S1 and S2). δ13C differed among regions for all species: spotted seatrout (F2,27 = 40.77, p < 0.001), black drum (F2,27 = 28.94, p < 0.001), bull shark (F2,27 = 9.89, p < 0.001), and alligator gar (F2,27 = 28.66, p < 0.001; Fig. 4a). For all species but bull shark, Trinity Bay and East Bay δ13C did not differ (all Tukey contrasts, p < 0.001) but were both lower than West Bay (spotted seatrout p = 0.074, black drum p = 0.513, and alligator gar p = 0.224). For bull sharks, δ13C was lowest in Trinity Bay (East Bay p = 0.002, West Bay p < 0.001), while East Bay and West Bay were similar (p = 0.605). δ15N also differed among regions for all species: spotted seatrout (F2,27 = 62.78, p < 0.001), black drum (F2,27 = 18.78, p < 0.001), bull shark (F2,27 = 12.81, p < 0.001), and alligator gar (F2,27 = 8.06, p = 0.002; Fig. 4b). For spotted seatrout, the highest δ15N values were observed in Trinity Bay, followed by East Bay and lastly West Bay (Tukey contrasts, all p < 0.001). Black drum δ15N was highest in Trinity Bay and East Bay (p = 0.537) and lowest in West Bay (both contrasts, p < 0.001). Bull shark δ15N values were highest in Trinity Bay (both contrasts, p < 0.001) and lowest in East Bay and West Bay (no difference, p = 0.864). Similar to black drum, alligator gar δ15N was highest in Trinity Bay and East Bay (p = 0.113) and lowest in West Bay (Trinity Bay p = 0.001, East Bay p = 0.036).
Boxplots of δ13C (a) and δ15N (b) by species in each region, with each species represented by colors as defined in the legend. Boxes represent the 25th percentile, median, and 75th percentile, with whiskers representing 1.5 times the interquartile range. Outliers (data points that fall outside the range of the whiskers) are represented by points
Source Contributions and Trophic Niche Overlap
Proportional source contributions differed by region for all species, but FW POM & Plants and Marine POM always contributed the greatest proportions (Fig. 5). Furthermore, contribution of FW POM & Plants was generally highest in regions with high freshwater influence (Trinity Bay, East Bay), while the opposite was true for Marine POM (Fig. 5). For spotted seatrout, the median contribution of FW POM & Plants and Marine POM was 48.6% and 41.1% in Trinity Bay, 43.6% and 45.0% in East Bay, and 35.5% and 49.7% in West Bay, respectively. For black drum, the median contribution of FW POM & Plants and Marine POM was similar between Trinity Bay (54.2% and 30.9%, respectively) and East Bay (56.1% and 30.1%, respectively), but in West Bay, there were greater contributions of Marine POM (39.2%) and Marine Plants (12.7%) and lower contribution of FW POM & Plants (32.4%). Of these species, bull sharks generally had the highest contribution of Marine Plants (7.4%, 14.1%, and 14.4% in Trinity, East, and West Bays). For bull sharks in Trinity Bay, FW POM & Plants and Marine POM contributed similarly (41.0% and 42.0%, respectively), but Marine POM contribution was higher than FW POM & Plants in East Bay (46.1% and 25.8%) and West Bay (47.3% and 25.6%). Alligator gar contributions most clearly followed a pattern of increasing marine influence and decreasing freshwater influence from Trinity Bay to West Bay; Freshwater POM & Plants and Marine POM contributions were 57.6% and 31.6% in Trinity Bay, 48.7% and 37.8% in East Bay, and 26.5% and 45.0% in West Bay, respectively. Marine Plants contributed 9.5% in West Bay. Unless otherwise noted, Marine Plants and FW Spartina had negligible contributions for all species in all regions (< 5%; Fig. 5).
Hypervolume-based trophic niche size and overlap differed among regions for each species, in some cases dramatically (Fig. 6). Spotted seatrout trophic niche increased from Trinity Bay (5.19) to East Bay (17.34) and was largest in West Bay (54.81; Table 3). For black drum, trophic niche size was largest in East Bay (107.02), followed by West Bay (61.48) and Trinity Bay (12.09). Bull sharks had the most notable difference in trophic niche size among regions: 1454.99 in Trinity Bay, 62.28 in East Bay, and 2.95 in West Bay. Oppositely, Alligator gar trophic niche was largest in West Bay (461.56), followed by East Bay (12.80) and Trinity Bay (5.24; Fig. 6 and Table 3). These spatial differences in trophic niche size, whereby a different species in each region had the largest niche, translated to regionally unique patterns of overlap (Fig. 6 and Table 4). In Trinity Bay, the large trophic niche of bull sharks encompassed all other species, resulting in low overlap indices (0.007 to 0.015) and high unique fractions for bull sharks (0.99 to 1). The other species exhibited relatively small and moderately overlapping (0.33 to 0.48) trophic niches. In East Bay, the large trophic niche of black drum exhibited low overlap with other species (0.085 to 0.17) and high unique fractions (0.87 to 0.95). Trophic niches of the other species were moderate in size and ranged from low (0.16) between alligator gar and bull sharks to moderate (0.43) between alligator gar and spotted seatrout. In West Bay, alligator gar had the largest trophic niche that had low overlap (0.013 to 0.19) with other species and high unique fractions (0.89 to 0.99). Overlap was moderate between spotted seatrout and black drum (0.38), but low between bull sharks and both spotted seatrout (0.07) and black drum (0.04; Fig. 6 and Table 4).
Trophic Position Estimation
In order from highest to lowest, bulk SIA TP estimates calculated using a pooled baseline were 4.03 ± 0.51 for spotted seatrout, 3.77 ± 0.76 for alligator gar, 3.69 ± 0.64 for bull shark, and 3.38 ± 0.64 for black drum (Table 5). There was some variation in TP estimates when regional baselines were used, but all regional estimates differed from the pooled estimate by < 0.6 (Table 5). Average regional TP estimates were 4.03 ± 0.33 for spotted seatrout, 3.81 ± 0.62 for alligator gar, 3.67 ± 0.51 for bull shark, and 3.39 ± 0.48 for black drum (Fig. 7). TP estimates calculated using CSIA-AA were 3.89 ± 0.16 for spotted seatrout, 3.87 ± 0.09 for bull shark, 3.75 ± 0.25 for alligator gar, and 3.13 ± 0.40 for black drum (Table 5 and Fig. 7). There was no interaction between method (regional bulk SIA vs. CSIA-AA) and species (F3,136 = 0.55, p = 0.48) and minimal difference between methods (F1,139 = 0.09, p = 0.52). A range of TP emerged among the species (F3,140 = 12.65, p < 0.001; Fig. 7), the lowest of which was black drum (Tukey contrasts, p < 0.001 for spotted seatrout and alligator gar, p = 0.003 for bull shark). Bull shark TP was lower than spotted seatrout (p = 0.02), but similar to alligator gar (p = 0.42). The species with the highest TP was spotted seatrout, followed closely by alligator gar (p = 0.06).
Ontogenetic Shifts in Trophic Position
The total length of some species differed among regions, confounding ecosystem-wide interpretation of ontogenetic shifts in TP due to regional differences in baseline δ15N. Spotted seatrout total length differed among regions (F2,27 = 4.15, p = 0.03), with larger individuals in West Bay (mean STL 471.8 mm) compared to Trinity Bay (393.5 mm, p = 0.03) and East Bay (412.1 mm, p = 0.045) but similar between Trinity Bay and East Bay (p = 0.52). Black drum total length also differed among regions (F2,27 = 72.00, p < 0.001), with the largest individuals in Trinity Bay (830.3 mm), followed by East Bay (715.1 mm) and lastly West Bay (428.5 mm, all contrasts p < 0.001). Bull shark total length differed among regions (F2,27 = 4.04, p = 0.03), with larger individuals captured in West Bay (1168.5 mm) compared to East Bay (975.3 mm, p = 0.02) but similar between Trinity Bay (1057.8 mm) and either East Bay (p = 0.23) or West Bay (p = 0.10). Alligator gar total length was similar among regions: Trinity Bay (1047.5 mm), East Bay (1046.2 mm), and West Bay (1085.2 mm, F2,27 = 0.24, p = 0.78).
Given the observed regional differences in total length, the relationship between bulk SIA TP estimates and total length was compared using pooled vs. regional baseline δ15N values (Fig. 8). For spotted seatrout, there was no apparent relationship between TP and total length when TP was calculated with a pooled baseline value (R2 = 0.05, n = 30, p = 0.22; Fig. 8a). However, there was a positive relationship when TP was calculated with the more appropriate regional baseline (R2 = 0.28, n = 30, p = 0.003; Fig. 8b). The opposite pattern was observed for black drum: pooled baseline TP estimates exhibited a positive relationship with total length (R2 = 0.49, n = 30, p < 0.001; Fig. 8c), while regional baseline TP estimates showed no relationship with total length (R2 = 0.08, n = 30, p = 0.14; Fig. 8d). Similar to spotted seatrout, bull shark pooled TP estimates exhibited a very weak relationship with total length (R2 = 0.12, n = 30, p = 0.06; Fig. 8e), but a strong positive relationship emerged between regional TP and total length (R2 = 0.46, n = 30, p < 0.001; Fig. 8f). Since alligator gar total length was consistent between regions, similar positive relationships were observed between pooled TP and total length (R2 = 0.26, n = 30, p = 0.004; Fig. 8g) and between regional TP and total length (R2 = 0.55, n = 30, p < 0.001; Fig. 8h).
Trophic position estimates by total length (mm), with a pooled baseline value (left panels) and regional baseline values (right panels) for each species: spotted seatrout (a, b), black drum (c, d), bull shark (e, f), and alligator gar (g, h). Linear regression line of best fit (black line), 95% confidence intervals (shaded gray), regression equation, and R2 values are displayed for statistically significant relationships only (p < 0.05)
Stomach Contents (Alligator Gar)
Of the 57 alligator gar stomachs examined, 32 contained prey items (56.1% full). Prey from three major taxa were found, with the vast majority of stomachs containing teleost fishes (93.8%FO) and very few containing insects (6.3%FO) or mammals (3.1%FO; Table 6). Of identifiable teleost fishes, the majority were of the genus Ariidae (15.6%FO) and Clupeidae (12.5%FO), with other genera including Mugilidae, Sciaenidae, Ophichthidae, and Gobiidae less represented (< 10%FO). Unidentifiable insect remains were present in two stomachs, and a small mammal of the order Rodentia was present in one.
Discussion
This study describes spatial and ontogenetic patterns in the trophic ecology of a model assemblage of estuarine predators and provides evidence for the importance of spatially explicit stable isotope analysis. The observed heterogeneity in both baseline and consumer stable isotope ratios indicates that assessing trophic ecology of estuarine consumers with SIA requires careful consideration of spatial scale. Specifically, baseline δ15N values were distinctly higher in regions characterized by low salinity (i.e., Trinity Bay and East Bay), which most likely reflects anthropogenically derived nitrogenous inputs from the main sources of freshwater to the GBC. Large freshwater pulses which decrease salinity in the GBC are correlated to rapid increases in concentrations of nitrate, nitrite, and ammonium (Steichen et al. 2020). However, the volume of freshwater input is usually small compared to the volume of the GBC, meaning the system has a fairly slow flushing rate (0.08 d−1; Roelke et al. 2013). Nitrogenous inputs are therefore unlikely to be removed quickly from the system via flushing, and our results indicate they are incorporated into the food web. Spatial patterns of baseline δ15N indeed translated to similar patterns in predator ratios, with δ15N values of all four species being more enriched in lower-salinity regions of the GBC. Similar patterns of shifting baseline δ15N values with salinity have been observed in other estuaries (McClelland et al. 1997; Jennings and Warr 2003; Bishop et al. 2017; Matich et al. 2021), further demonstrating that this phenomenon should be considered in any system that may be affected by nitrogenous inputs.
Source contributions derived from mixing models revealed spatially distinct patterns of basal resource use, with either marine or freshwater POM being the greatest contributor to all species’ food webs across regions. The observed reliance on pelagic basal resources aligns with known prey taxa consumed by these predators, with diets often consisting of filter-feeding or planktivorous fishes and invertebrates (e.g., Mugil spp., Brevoortia spp., and Penaeid shrimp; Hall-Scharf et al. 2016; Rubio et al. 2018; TinHan and Wells 2021). The proportional contribution of marine and freshwater POM generally followed a logical pattern along the estuarine salinity continuum, whereby freshwater POM and plants contributed the most in Trinity and East Bays (low salinity), while Marine POM contributed the most in West Bay (high salinity). Furthermore, proportional contribution of marine macrophytes (Spartina, seagrasses) was highest in West Bay for all species but spotted seatrout, suggesting an increased reliance on autochthonous benthic production in the low estuary where allochthonous (riverine/terrestrial) particulate matter from freshwater inflow are less dominant. The spatial arrangement of benthic primary producers also contributes to this pattern, since the generally high turbidity in Trinity and East Bays impedes the growth of submerged aquatic vegetation. Patterns of basal resource incorporation across estuarine gradients are complex and often depend significantly on the hydrological context of the system (Chanton and Lewis 2002; Nelson et al. 2015; Possamai et al. 2020). Our results indicate that regional differences in food web structure occur within the GBC, driven by patterns of freshwater inflow and the availability and distribution of basal resources.
Spatially distinct basal resource contributions translated to regional differences in trophic niche volume and shape for all predators, and species-specific patterns emerged that were likely mediated by their unique habitat use patterns, movements, and feeding behaviors. These species represent a model assemblage of predators in estuaries, ranging from highly euryhaline to freshwater associated, from piscivorous to benthivorous, and from highly to moderately mobile. We observed patterns of intraspecific niche expansion and contraction within the estuary that were mediated by habitat, likely resulting from a suite of intrinsic (e.g., movement) and extrinsic (e.g., resource availability) factors. Predator foraging movements in particular can be highly influential to shaping ecosystem structure and function by linking disparate food webs and transferring energy and nutrients across habitats (Polis et al. 1997; Rooney et al. 2008). This trophic coupling can result in spatial and temporal subsidies that act to stabilize food webs by dampening spatiotemporal fluctuations in prey populations (McCann et al. 2005). Our results suggest that these species may represent unique trophic typologies, with distinct movement and habitat use patterns that influence their trophic roles at the landscape scale, each contributing to the connectivity of habitats across the estuary.
Bull sharks exhibited the greatest difference in trophic niche among regions, with a three order of magnitude decrease in niche volume from the freshest (Trinity Bay) to most marine (West Bay) habitats. Bull sharks are highly mobile euryhaline predators, but juveniles use low-salinity estuarine habitats like Trinity Bay for nursery and refuge space (Heupel and Simpfendorfer 2011; Matich et al. 2020). Their expanded niche and similar contributions of marine and freshwater basal resources in Trinity Bay indicate that individuals in this optimal habitat may undergo roving movements to and from the low estuary to capitalize on feeding opportunities before returning. Individual variability in roving behavior has been observed for juvenile bull sharks in a South Florida, USA, estuary, with increased use of marine habitats at larger sizes (Matich and Heithaus 2015). Individuals captured in marine habitats (West Bay) were slightly larger than those in other regions, and their contracted niche and reliance on Marine POM suggests that they were feeding locally. Spatial and ontogenetic differences in roving behavior of juvenile bull sharks likely alter the degree of trophic coupling they provide across the estuarine landscape, with greater connection between fresh and marine habitats provided by small, roving individuals primarily residing in freshwater nursery habitats.
Despite having similar associations with low salinities, the observed spatial pattern of alligator gar trophic niche volume was opposite from that of bull sharks; trophic niche volume increased by two orders of magnitude from fresh to marine habitats. Alligator gar in Galveston Bay are primarily associated with low salinities but can be found throughout the estuary (Livernois et al. 2021). Their movements within estuarine environments are poorly resolved, but a study in the Trinity River described fairly small home ranges (< 60 km linear distance for 80% of individuals; Buckmeier et al. 2013). Given their general preference for low salinities and moderate degree of movement, the relatively small trophic niche observed in Trinity Bay suggests that they feed locally while inhabiting fresh and brackish habitats. The comparatively large niche observed in West Bay could be the result of large-scale roving behavior as suggested for bull sharks but may also be explained by the diversity of basal resources in that region. West Bay is hydrologically separated from the main sources of freshwater inflow to the GBC but does receive input from the Chocolate Bayou watershed (Guthrie et al. 2012). The expanded trophic niche of alligator gar in West Bay may not be a result of inter-region movements but rather local feeding on diverse prey via intra-region movements between marine and freshwater habitats. This suggests that alligator gar play a unique trophic role in the low estuary, potentially coupling fresh and marine habitats across a relatively small spatial scale.
Black drum differed from bull sharks and alligator gar in that their trophic niche volume was largest in East Bay, suggesting that individuals in the mid-estuary may forage between fresh and marine habitats, while contracted niches in Trinity and West Bays indicated local feeding. Adult black drum are extremely euryhaline and highly mobile, surviving in salinities from nearly 0 to 60+ ppt and traveling up to 80 km/day (Ajemian et al. 2018; Livernois et al. 2021). Their tolerance of a wide range of salinities and ability to move great distances may allow black drum to forage freely throughout habitats in the estuary, but our results only provide evidence of this for individuals captured in the mid-estuary. However, the individuals in this study differed in size among regions (largest in Trinity Bay, moderately sized in East Bay, and smallest in West Bay), and little is known about the movement and foraging patterns of black drum across subadult to adult life stages. Examining the movements of multiple size classes would help to disentangle the habitat- and size-specific roles black drum play in the trophic coupling of fresh, brackish, and marine habitats in estuaries.
Spotted seatrout, the smallest-bodied predator of this assemblage, exhibited the least notable differences in trophic niche volume among regions. Furthermore, the contribution of marine and freshwater basal resources closely matched the habitat in which they were captured, suggesting that they had been foraging locally (within tissue turnover time, ~ 1 to 2 months for a related species; Mont’Alverne et al. 2016). Spotted seatrout are euryhaline and are found throughout estuarine systems, and our results indicate that they feed locally within fresh, brackish, and marine environments. Spotted seatrout are able to move large distances in response to environmental forcing (e.g., freshets; Callihan et al. 2015), but preliminary assessment of their movements in Galveston Bay suggests relatively small core use areas (Livernois 2022). However, their habitat use patterns change seasonally, with increased use of up-estuary habitats (Trinity Bay) in the fall compared to spring (Livernois et al. 2021). Our results suggest that spotted seatrout may not provide extensive trophic coupling among habitats throughout the estuary within a given season but may do so across longer temporal scales as they move between habitats seasonally. An acoustic telemetry study of spotted seatrout, black drum, bull sharks, and alligator gar in Galveston Bay is ongoing, and those data will provide greater insight into the unique, spatiotemporally distinct trophic roles of each of these predators across the estuarine landscape.
Since each species’ trophic niche differed spatially, so too did the degree of niche overlap and partitioning between them. In each region, a single species exhibited a large trophic niche that encompassed the majority of niche space of the other species (e.g., bull sharks in Trinity Bay, black drum in East Bay, and alligator gar in West Bay). As a result, species with smaller trophic niches experienced very little unique niche space in that region. However, niche overlap was never greater than 0.5 between any combination of species in any region, suggesting a moderate degree of partitioning. This spatially distinct degree of niche overlap and partitioning among species highlights the unique role of each predator and provides evidence for mechanisms contributing to ecosystem stability. The observed partitioning of these species’ trophic niches may aid in allowing them to coexist within the same habitats (MacArthur 1958; Hutchinson 1959), while niche overlap among predators can protect the ecosystem from perturbation via trophic redundancy (Naeem 1998; Sanders et al. 2018; Biggs et al. 2020). However, despite some degree of niche overlap within a given region, our results suggest that these species are not redundant nor interchangeable. Each species exhibited unique patterns of niche expansion and contraction that were likely related to habitat use and movement patterns that impart trophic stability across the estuarine-coastal landscape (Chalcraft and Resetarits Jr. 2003; McCann et al. 2005; Rooney et al. 2008). Predator functional diversity can have strong effects on the structure and function of ecosystems (Schmitz 2009; Lefcheck and Duffy 2015), but it is often negatively influenced by anthropogenic pressures (e.g., fishing (Clemente et al. 2010) and habitat loss (Kissick et al. 2018)). Effective conservation of estuarine ecosystems therefore relies on careful, species-specific management of these predators, all of which are subject to recreational fishing pressure and habitat loss in Gulf of Mexico estuaries and coasts.
In addition to spatial differences in trophic ecology of these predators, ontogenetic (size-based) shifts in trophic position also emerged for all species but black drum whereby TP increased with body size. Many fish species exhibit increasing TP with size, but the strength and direction of these relationships often depend on functional traits and morphology (Keppeler et al. 2020). Size-based shifts in TP and stable isotope ratios occur for spotted seatrout and bull sharks, with ontogenetic dietary changes corresponding to a positive relationship between length and both TP and δ15N (Wenner and Archambault 1996; Werry et al. 2011; Fulford and Dillon 2013; Cottrant et al. 2021; Matich et al. 2021; TinHan and Wells 2021). Alligator gar diets and stable isotope ratios are not well resolved, but a study in an Oklahoma reservoir determined that prey size increased with alligator gar length (Snow and Porta 2020), which may explain the observed increase in TP with length in the present study. Adult black drum diet is similar between individuals of 200 to 400 mm TL (Rubio et al. 2018), and our results indicate that this remains true for larger individuals (400–1000 mm TL). For species that did exhibit ontogenetic shifts in TP, a difference of 1–2 trophic levels was observed between the smallest and largest individuals, significantly shifting the ecological role of each predator over time. TP estimates are often used in ecosystem models to examine food web structure and function (Deehr et al. 2014; Pethybridge et al. 2018), so it is important to examine TP across a range of sizes to accurately categorize the ecological role of a given species across life stages. Our results also highlight the importance of using spatially explicit baseline δ15N values in estuarine ecosystems, given the discrepancies observed between length-TP relationships when using pooled versus regional baseline values. Our results closely match those of Matich et al. (2021) in San Antonio Bay, Texas, approximately 200 km south of the GBC. A similar increase in baseline δ15N with decreasing salinity was observed, which was used to estimate TP of juvenile bull sharks using pooled and salinity-corrected baseline values. Our results corroborate the findings of Matich et al. (2021) and exemplify the importance of spatial scale when analyzing and interpreting stable isotope data in estuarine systems.
While stable isotope ratios exhibited distinct spatial patterns in the GBC, neither δ13C nor δ15N values differed temporally between the two seasons (spring and fall) for primary producers or predators. A previous study of the same species in the GBC noted seasonal shifts in their habitat use patterns and their degree of spatiotemporal overlap with putative prey, suggesting the potential for seasonal shifts in diet (Livernois et al. 2021). No evidence of this seasonality was observed in the present study using SIA. Stomach content analysis can provide a finer-scale assessment of diets compared to SIA (Richards et al. 2023), but our limited assessment of the gut contents of a single species (alligator gar) precluded seasonal analysis (Table 6). Seasonality in prey consumption has been observed for alligator gar in other Texas estuaries, but concurrent isotopic niche calculations were less seasonally distinct (Marsaly et al. 2023). It is therefore possible that the exact prey taxa being consumed by these predators may shift seasonally, but similarities in isotopic ratios among those prey mute the effect in predator isotopic niches.
Stomach contents of these predators have been fairly well resolved in estuaries, with the exception of alligator gar. They have been documented as primarily piscivorous in freshwater systems, with most gut content studies rarely reporting other taxa besides fish in their diet (García de León et al. 2001; Snow and Porta 2020). The diets of alligator gar in two Texas estuaries (north and south of the GBC) also contained almost exclusively fish, dominated by mugilids and clupeids (Marsaly et al. 2023). Our results expand upon this dietary knowledge of alligator gar in estuaries; their stomachs contained primarily fish (from at least 6 genera) but also previously unreported taxa including insects and a small mammal (a rodent, potentially Ondatra or Myocastor). Their generally piscivorous diet that includes the occasional odd prey taxa suggests a fairly flexible feeding strategy, which may partially explain the shift in trophic niche size among regions if prey diversity differs spatially. Further examination of the potential prey and diets of alligator gar, and the other species in this study, would provide stronger insight into potential trophic plasticity.
In conclusion, this study elucidated the spatial and ontogenetic trophic ecology of an assemblage of co-occurring estuarine predators. Spatially distinct trophic niche volumes, and therefore overlap, suggested that these species occupy unique trophic roles among regions throughout the estuary. The observed niche size variation was likely related to differences in habitat use patterns and movement ecology of each species, which may provide important trophic coupling across the freshwater-marine continuum. In addition to spatial complexity in each species’ trophic ecology, ontogenetic shifts in trophic position were observed for three species, altering their role as predators over their lifespan. Estuaries like the GBC are at the forefront of anthropogenic impacts such as excess nutrient inputs, habitat degradation, and fishery pressure (Lotze et al. 2006). Our results delineate the individual trophic roles of multiple estuarine predators and provide evidence for the importance of predator diversity in structuring ecosystems and food webs.
References
Ajemian, M.J., K.S. Mendenhall, J. Beseres Pollack, M.S. Wetz, and G.W. Stunz. 2018. Moving forward in a reverse estuary: Habitat use and movement patterns of black drum (Pogonias cromis) under distinct hydrological regimes. Estuaries and Coasts 41: 1410–1421. https://doi.org/10.1007/s12237-017-0363-6.
Allgeier, J.E., T.J. Cline, T.E. Walsworth, G. Wathen, C.A. Layman, and D.E. Schindler. 2020. Individual behavior drives ecosystem function and the impacts of harvest. Science Advances 6: eaax8329. https://doi.org/10.1126/sciadv.aax8329.
Biggs, C.R., L.A. Yeager, D.G. Bolser, C. Bonsell, A.M. Dichiera, Z. Hou, S.R. Keyser, A.J. Khursigara, K. Lu, A.F. Muth, B. Negrete Jr., and B.E. Erisman. 2020. Does functional redundancy affect ecological stability and resilience? A Review and Meta-Analysis. Ecosphere 11 (7): e03184. https://doi.org/10.1002/ecs2.3184.
Bishop, K.A., J.W. McClelland, and K.H. Dunton. 2017. Freshwater contributions and nitrogen sources in a south Texas estuarine ecosystem: A time-integrated perspective from stable isotopic ratios in the eastern oyster (Crassostrea virginica). Estuaries and Coasts 40 (5): 1314–1324. https://doi.org/10.1007/s12237-017-0227-0.
Blonder, B., C. Lamanna, C. Violle, and B.J. Enquist. 2014. The n-dimensional hypervolume. Global Ecology and Biogeography 23: 595–609. https://doi.org/10.1111/geb.12146.
Blonder, B., C.B. Morrow, S. Brown, G. Butruille, D. Chen, A. Laini, and D.J. Harris. 2023. Hypervolume: High dimensional geometry, set operations, projection, and inferences using kernel density estimation, support vector machines, and convex hulls. R Package Version 3 (1): 3.
Bolnick, D.I., R. Svanbäck, J.A. Fordyce, L.H. Yang, J.M. Davis, C.D. Hulsey, M.L. Forister, and M.A. McPeek. 2003. The ecology of individuals: Incidence and implications of individual specialization. The American Naturalist 161: 1–28. https://doi.org/10.1086/343878.
Bradley, C.J., N.J. Wallsgrove, A. Choy, J.C. Drazen, E.D. Hetherington, D.K. Hoen, and B.N. Popp. 2015. Trophic position estimates of marine teleosts using amino acid compound specific isotope analysis. Limnology and Oceanography: Methods 13: 476–493. https://doi.org/10.1002/lom3.10041.
Buckmeier, D.L., N.G. Smith, and D.J. Daugherty. 2013. Alligator gar movement and macrohabitat use in the lower Trinity River, Texas. Transactions of the American Fisheries Society 142: 1025–1035. https://doi.org/10.1080/00028487.2013.797494.
Callihan, J.L., J.H. Cowan Jr., and M.D. Harbison. 2015. Sex-specific movement response of an estuarine sciaenid (Cynoscion nebulosus) to freshets. Estuaries and Coasts 38: 1492–1504. https://doi.org/10.1007/s12237-014-9889-z.
Cardinale, B.J., J.E. Duffy, A. Gonzalez, D.U. Hooper, C. Perrings, P. Venail, A. Narwani, G.M. Mace, D. Tilman, D.A. Wardle, A.P. Kinzig, G.C. Daily, M. Loreau, J.B. Grace, A. Larigauderie, D.S. Srivastava, and S. Naeem. 2012. Biodiversity loss and its impact on humanity. Nature 486: 59–67. https://doi.org/10.1038/nature11148.
Chalcraft, D.R., and W.J. Resetarits Jr. 2003. Predator identity and ecological impacts: Functional redundancy or functional diversity? Ecology 84: 2407–2418. https://doi.org/10.1890/02-0550.
Chanton, J., and F.G. Lewis. 2002. Examination of coupling between primary and secondary production in a river-dominated estuary: Apalachicola Bay, Florida, U.S.A. Limnology and Oceanography 47: 683–697. https://doi.org/10.4319/lo.2002.47.3.0683.
Chikaraishi, Y., N.O. Ogawa, Y. Kashiyama, Y. Takano, H. Suga, A. Tomitani, H. Miyashita, H. Kitazato, and N. Ohkouchi. 2009. Determination of aquatic food-web structure based on compound-specific nitrogen isotopic composition of amino acids. Limnology and Oceanography: Methods 7 (11): 740–750. https://doi.org/10.4319/lom.2009.7.740.
Clemente, S., J.C. Hernández, A. Rodríguez, and A. Brito. 2010. Identifying keystone predators and the importance of preserving functional diversity in sublittoral rocky-bottom areas. Marine Ecology Progress Series 413: 55–67. https://doi.org/10.3354/meps08700.
Cloern, J.E., S.Q. Foster, and A.E. Kleckner. 2014. Phytoplankton primary production in the world’s estuarine-coastal ecosystems. Biogeosciences 11: 2477–2501. https://doi.org/10.5194/bg-11-2477-2014.
Cottrant, E., P. Matich, and M.R. Fisher. 2021. Boosted regression tree models predict the diets of juvenile bull sharks in a subtropical estuary. Marine Ecology Progress Series 659: 127–141. https://doi.org/10.3354/meps13568.
Deehr, R.A., J.J. Luczkovich, K.J. Hart, L.M. Clough, B.J. Johnson, and J.C. Johnson. 2014. Using stable isotope analysis to validate effective trophic levels from Ecopath models of areas closed and open to shrimp trawling in Core Sound, NC, USA. Ecological Modeling 282: 1–17. https://doi.org/10.1016/j.ecolmodel.2014.03.005.
DeNiro, M.J., and S. Epstein. 1978. Influence of diet on the distribution of carbon isotopes in animals. Geochimica Et Cosmochimica Acta 42: 495–506. https://doi.org/10.1016/0016-7037(78)90199-0.
Fox, J., and S. Weisberg. 2019. An R companion to applied regression, third edition. Thousand Oaks CA: Sage. https://socialsciences.mcmaster.ca/jfox/Books/Companion/
Fulford, R.S., and K. Dillon. 2013. Quantifying intrapopulation variability in stable isotope data for spotted seatrout (Cynoscion nebulosus). Fishery Bulletin 111 (2): 111–121. https://doi.org/10.7755/FB.111.2.1.
Galván, D.E., C.J. Sweeting, and W.D.K. Reid. 2010. Power of stable isotope techniques to detect size-based feeding in marine fishes. Marine Ecology Progress Series 407: 271–278. https://doi.org/10.3354/meps08528.
García de León, F.J., L. González-García, J.M. Herrera-Castillo, K.O. Winemiller, and A. Banda-Valdés. 2001. Ecology of the alligator gar, Atractosteus spatula, in the Vicente Guerrero Reservoir, Tamaulipas, México. The Southwestern Naturalist 46 (2): 151–157. https://doi.org/10.2307/3672523.
Gloeckler, K., C.A. Choy, C.C.S. Hannides, H.G. Close, E. Goetze, B.N. Popp, and J.C. Drazen. 2018. Stable isotope analysis of micronekton around Hawaii reveals suspended particles are an important nutritional source in the lower mesopelagic and upper bathypelagic zones. Limnology and Oceanography 63 (3): 1168–1180. https://doi.org/10.1002/lno.10762.
Grey, J., R.I. Jones, and D. Sleep. 2001. Seasonal changes in the importance of the source of organic matter to the diet of zooplankton in Loch Ness, as indicated by stable isotope analysis. Limnology and Oceanography 46: 505–513. https://doi.org/10.4319/lo.2001.46.3.0505.
Grubbs, R.D. 2010. Ontogenetic shifts in movements and habitat use. In: Carrier, J. C., Musick, J. A., and Heithaus, M. R. (eds) Sharks and their relatives. II. Biodiversity,adaptive physiology, and conservation. CRC, Boca Raton, pp 319–350. https://www.taylorfrancis.com/chapters/edit/10.1201/9781420080483-13/ontogenetic-shiftsmovements-habitat-use-dean-grubbs.
Guthrie, C.G., J. Matsumoto, and R.S. Solis. 2012. Analysis of the influence of water plan strategies on inflows and salinity in Galveston Bay. In Final Report to the United States Army Corps of Engineers, Contract #R0100010015. (Austin, TX: Texas Water Development Board), 71. https://www.twdb.texas.gov/surfacewater/bays/major_estuaries/trinity_san_jacinto/doc/WAMS_InfluenceGalBay_Final_20120822.pdf.
Hall-Scharf, B.J., T.S. Switzer, and C.D. Stallings. 2016. Ontogenetic and long-term diet shifts of a generalist juvenile predatory fish in an urban estuary undergoing dramatic changes in habitat availability. Transactions of the American Fisheries Society 145: 502–520. https://doi.org/10.1080/00028487.2016.1143396.
Hartman, K.J., and S.B. Brandt. 1995. Trophic resource partitioning, diets, and growth of sympatric estuarine predators. Transactions of the American Fisheries Society 124 (4): 520–537. https://doi.org/10.1577/1548-8659(1995)124%3c0520:TRPDAG%3e2.3.CO;2.
Hetherington, E.D., J.A. Seminoff, P.H. Dutton, L.C. Robison, B.N. Popp, and C.M. Kurle. 2018. Long-term trends in the foraging ecology and habitat use of an endangered species: An isotopic perspective. Oecologia 188: 1273–1285. https://doi.org/10.1007/s00442-018-4279-z.
Hette-Tronquart, N. 2019. Isotopic niche is not equal to trophic niche. Ecology Letters 22: 1987–1989. https://doi.org/10.1111/ele.13218.
Heupel, M.R., and C.A. Simpfendorfer. 2011. Estuarine nursery areas provide a low-mortality environment for young bull sharks Carcharhinus leucas. Marine Ecology Progress Series 433: 237–244. https://doi.org/10.3354/meps09191.
Hothorn, T., F. Bretz, and P. Westfall. 2008. Simultaneous inference in general parametric models. Biometrical Journal 50 (3): 346–363. https://doi.org/10.1002/bimj.200810425.
Hutchinson, G.E. 1959. Homage to Santa Rosalia or why are there so many kinds of animals? The American Naturalist 93: 145–159. https://doi.org/10.1086/282070.
James, W.R., J.S. Lesser, S.Y. Litvin, and J.A. Nelson. 2020. Assessment of food web recovery following restoration using resource niche metrics. Science of the Total Environment 711: 134801. https://doi.org/10.1016/j.scitotenv.2019.134801.
Jennings, S., and K.J. Warr. 2003. Environmental correlates of large-scale spatial variation in the δ15N of marine animals. Marine Biology 142: 1131–1140. https://doi.org/10.1007/s00227-003-1020-0.
Keppeler, F.W., C.G. Montaña, and K.O. Winemiller. 2020. The relationship between trophic level and body size in fishes depends on functional traits. Ecological Monographs 90 (4): e01415. https://doi.org/10.1002/ecm.1415.
Kim, S.L., and P.L. Koch. 2012. Methods to collect, preserve, and prepare elasmobranch tissues for stable isotope analysis. Environmental Biology of Fishes 95: 53–63. https://doi.org/10.1007/s10641-011-9860-9.
Kissick, A.L., J.B. Dunning Jr., E. Fernandez-Juricic, and J.D. Holland. 2018. Different responses of predator and prey functional diversity to fragmentation. Ecological Applications 28: 1853–1866. https://doi.org/10.1002/eap.1780.
Kroetz, A.M., J.M. Drymon, and S.P. Powers. 2016. Comparative dietary diversity and trophic ecology of two estuarine mesopredators. Estuaries and Coasts 40: 1171–1182. https://doi.org/10.1007/s12237016-0188-8.
Layman, C.A., M.S. Araujo, R. Boucek, C.M. Hammerschlag-Peyer, E. Harrison, Z.R. Jud, P. Matich, A.E. Rosenblatt, J.J. Vaudo, L.A. Yeager, D.M. Post, and S. Bearhop. 2012. Applying stable isotopes to examine food-web structure: An overview of analytical tools. Biological Reviews 87: 542–562. https://doi.org/10.1111/j.1469-185X.2011.00208.x.
Lefcheck, J.S., and J.E. Duffy. 2015. Multitrophic functional diversity predicts ecosystem functioning in experimental assemblages of estuarine consumers. Ecology 96: 2973–2983. https://doi.org/10.1890/14-1977.1.
Livernois, M.C., M. Fujiwara, M. Fisher, and R.J.D. Wells. 2021. Seasonal patterns of habitat suitability and spatiotemporal overlap within an assemblage of estuarine predators and prey. Marine Ecology Progress Series 668: 39–55. https://doi.org/10.3354/meps13700.
Livernois, M.C. 2022. Ecological dynamics and connectivity within an assemblage of predatory fishes in coastal Texas. Doctoral dissertation, Texas A&M University. https://oaktrust.library.tamu.edu/handle/1969.1/198049.
Loreau, M. 2004. Does functional redundancy exist? Oikos 104: 606–611. https://doi.org/10.1111/j.0030-1299.2004.12685.x.
Lotze, H.K., H.S. Lenihan, B.J. Bourque, R.H. Bradbury, R.G. Cooke, M.C. Kay, S.M. Kidwell, M.X. Kirby, C.H. Peterson, and J.B.C. Jackson. 2006. Depletion, degradation, and recovery potential of estuaries and coastal seas. Science 312: 1806–1809. https://doi.org/10.1126/science.1128035.
MacArthur, R.H. 1958. Population ecology of some warblers of northeastern coniferous forests. Ecology 39 (4): 599–619. https://doi.org/10.2307/1931600.
Marsaly, B., D. Daugherty, O. Shipley, C. Gelpi, N. Boyd, J. Davis, M. Fisher, and P. Matich. 2023. Contrasting ecological roles and flexible trophic interactions of two estuarine apex predators in the western Gulf of Mexico. Marine Ecology Progress Series 709: 55–76. https://doi.org/10.3354/meps14281.
Martinez-Andrade, M. Fisher, B. Bowling, and B. Balboa. 2009. Marine resource monitoring operations manual. Texas Parks and Wildlife Department Coastal Fisheries Division.
Matich, P., and M.R. Heithaus. 2015. Individual variation in ontogenetic niche shifts in habitat use and movement patterns of a large estuarine predator (Carcharhinus leucas). Oecologia 178: 347–359. https://doi.org/10.1007/s00442-015-3253-2.
Matich, P., M.R. Heithaus, and C.A. Layman. 2011. Contrasting patterns of individual specialization and trophic coupling in two marine apex predators. Journal of Animal Ecology 80 (1): 294–305. https://doi.org/10.1111/j.1365-2656.2010.01753.x.
Matich, P., R.J. Nowicki, J. Davis, J.A. Mohan, J.D. Plumlee, B.A. Strickland, T.C. TinHan, R.J.D. Wells, and M. Fisher. 2020. Does proximity to freshwater refuge affect the size structure of an estuarine predator (Carcharhinus leucas) in the north-western Gulf of Mexico? Marine and Freshwater Research 71: 1501–1516. https://doi.org/10.1071/MF19346.
Matich, P., O.N. Shipley, and O.C. Weideli. 2021. Quantifying spatial variation in isotopic baselines reveals size-based feeding in a model estuarine predator: Implications for trophic studies in dynamic ecotones. Marine Biology 168: 108. https://doi.org/10.1007/s00227-021-03920-0.
McCann, K.S., J.B. Rasmussen, and J. Umbanhowar. 2005. The dynamics of spatially coupled food webs. Ecology Letters 8: 523–523. https://doi.org/10.1111/j.1461-0248.2005.00742.x.
McClelland, J.W., and J.P. Montoya. 2002. Trophic relationships and the nitrogen isotopic composition of amino acids in plankton. Ecology 83 (8): 2173–2180. https://doi.org/10.1890/0012-9658(2002)083[2173:TRATNI]2.0.CO;2.
McClelland, J.W., I. Valiela, and R.H. Michener. 1997. Nitrogen-stable isotope signatures in estuarine food webs: A record of increasing urbanization in coastal watersheds. Limnology and Oceanography 42 (5): 930–937. https://doi.org/10.4319/lo.1997.42.5.0930.
McCutchan, J.H., Jr., W.M. Lewis Jr., C. Kendall, and C.C. McGrath. 2003. Variation in trophic shift for stable isotope ratios of carbon, nitrogen, and sulfur. Oikos 102: 378–390. https://doi.org/10.1034/j.1600-0706.2003.12098.x.
McMahon, K.W., L.L. Hamady, and S.R. Thorrold. 2013. A review of ecogeochemistry approaches to estimating movements of marine animals. Limnology and Oceanography 58: 697–714. https://doi.org/10.4319/lo.2013.58.2.0697.
Minagawa, M., and E. Wada. 1984. Stepwise enrichment of 15N along food chains: Further evidence and the relation between δ15N and animal age. Geochimica et Cosmochimica Acta 48: 1135–1140. https://doi.org/10.1016/0016-7037(84)90204-7.
Mont’Alverne, R., Jardine, T. D., Pereya, P. E. R., Oliveira, M. C. L. M., Medeiros, R. S., Sampaio, L. A., Tesser, M. B., and Garcia, A. M. 2016. Elemental turnover rates and isotopic discrimination in a euryhaline fish reared under different salinities: Implications for movement studies. Journal of Experimental Marine Biology and Ecology 480: 36–44. https://doi.org/10.1016/j.jembe.2016.03.021.
Naeem, S. 1998. Species redundancy and ecosystem reliability. Conservation Biology 12 (1): 39–45. https://doi.org/10.1111/j.1523-1739.1998.96379.x.
Nelson, J.A., L. Deegan, and R. Garritt. 2015. Drivers of spatial and temporal variability in estuarine food webs. Marine Ecology Progress Series 533: 67–77. https://doi.org/10.3354/meps11389.
Newsome, S.D., J.D. Yeakel, P.V. Wheatley, and M.T. Tinker. 2012. Tools for quantifying isotopic niches pace and dietary variation at the individual and population level. Journal of Mammalogy 93: 329–341. https://doi.org/10.1644/11-MAMM-S-187.1.
Nielsen, J.M., B.N. Popp, and M. Winder. 2015. Meta-analysis of amino acid stable nitrogen isotope ratios for estimating trophic position in marine organisms. Oecologia 178: 631–642. https://doi.org/10.1007/s00442-015-3305-7.
Parnell, A.C., R. Inger, S. Bearhop, and A.L. Jackson. 2010. Source partitioning using stable isotopes: Coping with too much variation. PLoS ONE 5: e9672. https://doi.org/10.1371/journal.pone.0009672.
Pawluk, M., M. Fujiwara, and F. Martinez-Andrade. 2021. Climate effects on fish diversity in the subtropical bays of Texas. Estuarine, Coastal and Shelf Science 249: 107121. https://doi.org/10.1016/j.ecss.2020.107121.
Peterson, B.J., and B. Fry. 1987. Stable isotopes in ecosystem studies. Annual Review of Ecology and Systematics 18: 293–320. https://doi.org/10.1146/annurev.es.18.110187.001453.
Pethybridge, H.R., C.A. Choy, J.J. Polovina, and E.A. Fulton. 2018. Improving marine ecosystem models with biochemical tracers. Annual Review of Marine Science 10: 199–228. https://doi.org/10.1146/annurev-marine-121916-063256.
Plumlee, J.D., D.N. Hala, J.R. Rooker, J.B. Shipley, and R.J.D. Wells. 2021. Trophic ecology of fishes associated with artificial reefs assessed using multiple biomarkers. Hydrobiologia 848: 4347–4362. https://doi.org/10.1007/s10750-021-04647-1.
Polis, G.A., W.B. Anderson, and R.D. Holt. 1997. Toward an integration of landscape and food web ecology: The dynamics of spatially subsidized food webs. Annual Review of Ecology, Evolution and Systematics 28: 289–316. https://doi.org/10.1146/annurev.ecolsys.28.1.289.
Popp, B.N., B.S. Graham, R.J. Olson, C.C.S. Hannides, M.J. Lott, G.A. López-Ibarra, F. Galván-Magaña, and B. Fry. 2007. Insight into the trophic ecology of yellowfin tuna, Thunnus albacares, from Compound-Specific Nitrogen Isotope Analysis of Proteinaceous Amino Acids. Terrestrial Ecology 1: 173–190. https://doi.org/10.1016/S1936-7961(07)01012-3.
Possamai, B., D.J. Hoeinghaus, C. Odebrecht, P.C. Abreu, L.E. Moraes, A.C.A. Santos, and A.M. Garcia. 2020. Freshwater inflow variability affects the relative importance of allochthonous sources for estuarine fishes. Estuaries and Coasts 43: 880–893. https://doi.org/10.1007/s12237-019-00693-0.
Post, D.M. 2002. Using stable isotopes to estimate trophic position: Models, methods, and assumptions. Ecology 83 (3): 703–718. https://doi.org/10.1890/0012-9658(2002)083[0703:USITET]2.0.CO;2.
Post, D.M., C.A. Layman, D.A. Arrington, G. Takimoto, J. Quattrochi, and C.G. Montaña. 2007. Getting to the fat of the matter: Models, methods and assumptions for dealing with lipids in stable isotope analyses. Oecologia 152: 179–189. https://doi.org/10.1007/s00442-006-0630-x.
Powell, E.N., J.M. Klinck, E.E. Hofman, and M.A. McManus. 2003. Influence of water allocation and freshwater inflow on oyster production: A hydrodynamic–oyster population model for Galveston Bay, Texas, USA. Environmental Management 31 (1): 100–121. https://doi.org/10.1007/s00267-002-2695-6.
R Core Team. 2022. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. https://www.R-project.org/.
Rezek, R.J., J.A. Massie, J.A. Nelson, R.O. Santos, N.M. Viadero, R.E. Boucek, and J.S. Rehage. 2020. Individual consumer movement mediates food web coupling across a coastal ecosystem. Ecosphere 11: e03305. https://doi.org/10.1002/ecs2.3035.
Richards, T.M., T.T. Sutton, M.S. Woodstock, H. Judkins, and R.J.D. Wells. 2023. Body size, depth of occurrence, and local oceanography shape trophic structure in a diverse deep-pelagic micronekton assemblage. Progress in Oceanography 213: 102998. https://doi.org/10.1016/j.pocean.2023.102998.
Roelke, D.L., H. Li, N.J. Hayden, C.J. Miller, S.E. Davis, A. Quigg, and Y. Buyukates. 2013. Co-occurring and opposing freshwater inflow effects on phytoplankton biomass, productivity and community composition of Galveston Bay, USA. Marine Ecology Progress Series 477: 61–76. https://doi.org/10.3354/meps10182.
Rooney, N., K. McCann, G. Gellner, and J.C. Moore. 2006. Structural asymmetry and the stability of diverse food webs. Nature 442: 265–269. https://doi.org/10.1038/nature04887.
Rooney, N., K.S. McCann, and J.C. Moore. 2008. A landscape theory for food web architecture. Ecology Letters 11: 867–881. https://doi.org/10.1111/j.1461-0248.2008.01193.x.
Rubio, K.S., M. Ajemian, G.W. Stunz, T.A. Palmer, B. Lebreton, and J. Beseres Pollack. 2018. Dietary composition of black drum Pogonias cromis in a hypersaline estuary reflects water quality and prey availability. Journal of Fish Biology 93: 250–262. https://doi.org/10.1111/jfb.13654.
Sanders, D., E. Thébault, R. Kehoe, and F.J.F. van Veen. 2018. Trophic redundancy reduces vulnerability to extinction cascades. Proceedings of the National Academy of Sciences of the United States of America 115 (10): 2419–2424. https://doi.org/10.1073/pnas.1716825115.
Schmitz, O.J. 2009. Effects of predator functional diversity on grassland ecosystem function. Ecology 90: 2339–2345. https://doi.org/10.1890/08-1919.1.
Semmens, B.X., B.C. Stock, E. Ward, J.W. Moore, A. Parnell, A.L. Jackson, D.L. Phillips, S. Bearhop, and R. Inger. 2013. MixSIAR: A Bayesian stable isotope mixing model for characterizing intrapopulation niche variation. Minneapolis, Minnesota, USA: Ecological Society of America.
Shipley, O.N., and P. Matich. 2020. Studying animal niches using bulk stable isotope ratios: An updated synthesis. Oecologia 193: 27–51. https://doi.org/10.1007/s00442-020-04654-4.
Simonsen, K.A., and J.H. Cowan. 2013. Effects of an inshore artificial reef on the trophic dynamics of three species of estuarine fish. Bulletin of Marine Science 89 (3): 657–676. https://doi.org/10.5343/bms.2012.1013.
Snow, R.A., and M.J. Porta. 2020. Seasonal food habits and prey selectivity of alligator gar from Texoma Reservoir, Oklahoma. Journal of the Southeastern Association of Fish and Wildlife Agencies 8: 15–22.
Steichen, J.L., J.M. Labonté, R. Windham, D. Hala, K. Kaiser, S. Setta, P.C. Faulkner, H. Bacosa, G. Yan, M. Kamalanathan, and A. Quigg. 2020. Microbial, physical, and chemical changes in Galveston Bay following an extreme flooding event, hurricane Harvey. Frontiers in Marine Science 7: 186. https://doi.org/10.3389/fmars.2020.00186.
Stock, B.C., and B.X. Semmens. 2016. Unifying error structures in commonly used biotracer mixing models. Ecology 97: 2562–2569. https://doi.org/10.1002/ecy.1517.
Stock, B.C., A.L. Jackson, E.J. Ward, A.C. Parnell, D.L. Phillips, and B.X. Semmens. 2018. Analyzing mixing systems using a new generation of Bayesian tracer mixing models. PeerJ 6: e5096. https://doi.org/10.7717/peerj.5096.
TinHan, T.C., and R.J.D. Wells. 2021. Spatial and ontogenetic patterns in the trophic ecology of juvenile bull sharks (Carcharhinus leucas) from the northwest Gulf of Mexico. Frontiers in Marine Science 8: 664316. https://doi.org/10.3389/fmars.2021.664316.
Walker, B.H. 1992. Biodiversity and ecological redundancy. Conservation Biology 6 (1): 18–23. https://doi.org/10.1046/j.1523-1739.1992.610018.x.
Ward, E.J., B.X. Semmens, and D.E. Schindler. 2010. Including source uncertainty and prior information in the analysis of stable isotope mixing models. Environmental Science and Technology 44: 4645–4650. https://doi.org/10.1021/es100053v.
Wenner, C., and J. Archambault. 1996. Spotted seatrout natural history and fishing techniques in South Carolina. South Carolina Department of Natural Resources, Charleston. https://dc.statelibrary.sc.gov/server/api/core/bitstreams/5e4d287f-cba3-4d56-a2f0-81880e375db8/content.
Werry, J.M., S.Y. Lee, N.M. Otway, Y. Hu, and W. Sumpton. 2011. A multi-faceted approach for quantifying the estuarine–nearshore transition in the life cycle of the bull shark. Carcharhinus Leucas. Marine and Freshwater Research 62 (12): 1421–1431. https://doi.org/10.1071/MF11136.
Wilson, R.M., R.B. Tyson, J.A. Nelson, B.C. Balmer, J.P. Chanton, and D.P. Nowacek. 2017. Niche differentiation and prey selectivity among common bottlenose dolphins (Tursiops truncatus) sighted in St. George Sound, Gulf of Mexico. Frontiers in Marine Science 4: 235. https://doi.org/10.3389/fmars.2017.00235.
Woodcock, P., D.P. Edwards, R.J. Newton, F.A. Edwards, C.V. Khen, S.H. Bottrell, and K.C. Hamer. 2012. Assessing trophic position from nitrogen isotope ratios: Effective calibration against spatially varying baselines. Naturwissenschaften 99 (4): 275–283. https://doi.org/10.1007/s00114-012-0896-2.
Yarnes, C.T., and J. Herszage. 2017. The relative influence of derivatization and normalization procedures on the compound−specific stable isotope analysis of nitrogen in amino acids. Rapid Communications in Mass Spectrometry 31: 693–704. https://doi.org/10.1002/rcm.7832.
Acknowledgements
We thank the many TPWD staff members who aided in collecting samples for this project, including Christine Jensen, Kimberly Jeffrey, Derek York, Christopher Steffen, and Jared Cullison. We thank the students at Texas A&M University at Galveston for insightful discussions and assistance with field and lab work: Liam Batchelder, Emily Meese, Dr. Travis Richards, Dr. Tom TinHan, Annemarie Fougerousse, Anna DeMotte, Dr. Janelle Goeke, Shane Stephens, and Asif Mortuza. Thanks are due to Dr. Anna Armitage for the assistance with equipment needs and troubleshooting.
Funding
Open access funding provided by the Carolinas Consortium.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of Interest
The authors declare no competing interests.
Additional information
Communicated by James Allen Nelson
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Livernois, M.C., Rezek, R.J. & Wells, R.J.D. Spatial and Ontogenetic Trophic Dynamics of Co-occurring Predatory Fishes in a Northern Gulf of Mexico Estuary. Estuaries and Coasts 47, 1119–1138 (2024). https://doi.org/10.1007/s12237-024-01361-8
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12237-024-01361-8