Abstract
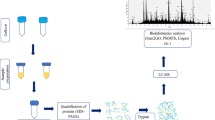
Our aim in this study was to characterize and investigate the secretome of Paenibacillus sp. S-12 by nanoLC-MS/MS tool-based analysis of trypsin digested culture supernatant proteins. Using a bioinformatics and combined approach of mass spectrometry, we identified 657 proteins in the secretome. Bioinformatic tools such as PREDLIPO, SecretomeP 2.0, SignalP 4.1, and PSORTb were used for the subcellular localization and categorization of secretome on basis of signal peptides. Among the identified proteins, more than 25% of the secretome proteins were associated with virulence proteins including flagellar, adherence, and immune modulators. Gene ontology analysis using Blast2GO tools categorized 60 proteins of the secretome into biological processes, cellular components, and molecular functions. KEGG pathway analysis identified the enzymes or proteins involved in various biosynthesis and degradation pathways. Functional analysis of secretomes reveals a large number of proteins involved in the uptake and exchange of nutrients, colonization, and chemotaxis. A good number of proteins were involved in survival and defense mechanism against oxidative stress, the production of toxins and antimicrobial compounds. The present study is the first report of the in-depth protein profiling of Paenibacillus bacterium. In summary, the current findings of Paenibacillus sp. S-12 secretome provide basic information to understand its survival and the possible pathogenic mechanism.





Similar content being viewed by others
Data availability
The data that support the findings of this study are in this published article and available from the corresponding author upon reasonable request.
References
Acosta H, Burchmore R, Naula C, Gualdrón-López M, Quintero-Troconis E, Cáceres AJ et al (2019) Proteomic analysis of glycosomes from Trypanosoma cruzi epimastigotes. Mol Biochem Parasitol 229:62–74
Aleti G, Sessitsch A, Brader G (2015) Genome mining: prediction of lipopeptides and polyketides from Bacillus and related Firmicutes. Comput Struct Biotechnol J 13:192–203
Bao K, Bostanci N, Selevsek N, Thurnheer T, Belibasakis GN (2015) Quantitative proteomics reveal distinct protein regulations caused by Aggregatibacter actinomycetemcomitans within sub-gingival biofilms. PLoS ONE 10:e0119222
Bao K, Bostanci N, Thurnheer T, Belibasakis GN (2017) Proteomic shifts in multi-species oral biofilms caused by Anaeroglobus geminatus. Sci Rep 7:1–11
Butler SM, Festa RA, Pearce MJ, Darwin KH (2006) Self-compartmentalized bacterial proteases and pathogenesis. Mol Microbiol 60:553–562
Capra EJ, Perchuk BS, Skerker JM, Laub MT (2012) Adaptive mutations that prevent crosstalk enable the expansion of paralogous signaling protein families. Cell 150:222–232
Carvalho APDA, Ventura GMC, Pereira CB, Leão RS, Folescu TW, Higa L, Teixeira LM, Maciel Plotkowski MC, Merquior VLC, Albano RM, Marques EA (2007) Burkholderia cenocepacia, B. multivorans, B. ambifaria and B. vietnamiensis isolates from cystic fibrosis patients have different profiles of exoenzyme production. APMIS 115:311–318
Cezairliyan B, Ausubel FM (2017) Investment in secreted enzymes during nutrient-limited growth is utility dependent. Proc Natl Acad Sci 114:E7796–E7802
Chevance FF, Hughes KT (2008) Coordinating assembly of a bacterial macromolecular machine. Nat Rev Microbiol 6:455–465
Clermont D, Gomard M, Hamon S, Bonne I, Fernandez JC, Wheeler R, Malosse C, Chamotrooke J, Gribaldo S, Gomperts BI (2015) Paenibacillus faecis sp. nov., isolated from human faeces. Int J Syst Evol Microl 65:4621
Conesa A, Gotz S, Garcia-Gomez JM, Terol J, Talon M, Robles M (2005) Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 21:3674–3676
Danese PN, Silhavy TJ (1998) Targeting and assembly of periplasmic and outer-membrane proteins in Escherichia coli. Annu Rev Genet 32:59–94
Ding R, Wu XC, Qian CD, Teng Y, Li O, Zhan ZJ, Zhao YH (2011) Isolation and identification of lipopeptide antibiotics from Paenibacillus elgii B69 with inhibitory activity against methicillin-resistant Staphylococcus aureus. J Microbiol 49:942–949
Elo S, Suominen I, Kampfer P, Juhanoja J, Salkinoja Salonen M, Haahtela K (2001) Paenibacillus borealis sp. nov., a nitrogen fixing species isolated from spruce forest humus in Finland. Int J Syst Evol Microbiol 51(2):535–545
Erhardt M, Namba K, Hughes KT (2010) Bacterial nanomachines: the flagellum and type III injectisome. Cold Sprin Harb Perspect Biol 2:a000299
Gagic D, Ciric M, Wen WX, Ng F, Rakonjac J (2016) Exploring the secretomes of microbes and microbial communities using filamentous phage display. Front Microbiol 7:429
Gagic D, Wen W, Collett MA, Rakonjac J (2013) Unique secreted-surface protein complex of Lactobacillus rhamnosus, identified by phage display. Microbiology Open 2:1–17
Ghio S, Ontañon O, Piccinni FE et al (2018) Paenibacillus sp. A59 GH10 and GH11 extracellular endoxylanases: application in biomass bioconversion. Bioenerg Res 11:174–190
Goosens VJ, Monteferrante CG, van Dijl JM (2014) The Tat system of Gram-positive bacteria. Biochem Biophys Acta 1843:1698–1706
Goulhen F, Hafezi A, Uitto VJ, Hinoda D, Nakamura R, Grenier D, Mayrand D (1998) Subcellular localization and cytotoxic activity of the GroEL-like protein isolated from Actinobacter actinomycetemcomitans. Infect Immun 66:5207–5213
Grady EN, MacDonald J, Liu L, Richman A, Yuan ZC (2016) Current knowledge and perspectives of Paenibacillus: a review. Microb Cell Fact 15:203
Guo L, Wu T, Hu W et al (2013) Phenotypic characterization of the foldase homologue PrsA in Streptococcus mutans. Mol Oral Microbiol 28:154–165
Haiko J, Westerlund-Wikström B (2013) The role of the bacterial flagellum in adhesion and virulence. Biology (Basel) 2:1242–1267
Hardoim PR, van Overbeek LS, van Elsas JD (2008) Properties of bacterial endophytes and their proposed role in plant growth. Trend Microbiol 16:463–471
Hemala L, Zhanga D, Margesin R (2014) Cold-active antibacterial and antifungal activities and antibiotic resistance of bacteria isolated from an alpine hydrocarbon contaminated industrialsite. Res Microbiol 165:447–456
Henderson B, Allan E, Coates AR (2006) Stress wars: the direct role of host and bacterial molecular chaperones in bacterial infection. Infect Immun 74:3693–4370
Hosogi Y, Duncan MJ (2005) Gene expression in Porphyromonas gingivalis after contact with human epithelial cells. Infect Immun 73:2327–2335
Hueck CJ (1998) Type III protein secretion systems in bacterial pathogens of animals and plants. Microbiol Mol Biol Rev 62:379–433
Hyyryläinen HL, Marciniak BC, Dahncke K, Pietiäinen M, Courtin P, Vitikainen M, Seppala R, Otto A, Becher D, Chapot-Chartier MP, Kuipers OP, Kontinen VP (2010) Penicillin-binding protein folding is dependent on the PrsA peptidyl-prolyl cis-trans isomerase in Bacillus subtilis. Mol Microbiol 77(1):108–127
Kim YS, Kotnala B, Kim YH, Jeon Y (2016) Biological characteristics of Paenibacillus polymyxa GBR-1 involved in root rot of stored Korean ginseng. J Ginseng Res 40:453–461
Kishore KH, Begum Z, Pathan AA, Shivaji S (2010) Paenibacillus glacialos sp. nov., isolated from Kafni Glacier of Himalayas. India Int J Syst Evol Microbiol 60:1909–1913
Lakshman DK, Natarajan SS, Lakshman S, Garrett WM, Dhar AK (2008) Optimized protein extraction methods for proteomic analysis of Rhizoctonia solani. Mycologia 100:867–875
Lee VT, Schneewind O (2001) Protein secretion and the pathogenesis of bacterial infections. Genes Dev 15:1725–1752
Lee W, Kwon D, Choi W, Jung GY, Au AK, Folch A, Jeon S (2015) 3D-printed microfluidic device for the detection of pathogenic bacteria using size-based separation in helical channel with trapezoid cross-section. Sci Rep 5:1–7
Lim JM, Jeon CO, Lee JC, Xu LH, Jiang CL, Kim CJ (2006) Paenibacillus gansuensis sp. nov., isolated from desert soil of Gansu Province in China. Int J Syst Evol Microbiol 56:2131–2134
Liu Y, Ding S, Shen J, Zhu K (2019) Non-ribosomal antibacterial peptides that target multidrug-resistant bacteria. Nat Prod Rep 36:573–592
Monciardini P, Iorio M, Maffioli S, Sosio M, Donadio S (2014) Discovering new bioactive molecules from microbial sources. Microbiol Biotechnol 7:209–220
Montes JM, Mercade´ E, Bozal N, Guinea J (2004) Paenibacillus antarcticus sp. nov., a novel psychrotolerant organism from the Antarctic environment. Int J Syst Evol Microbiol 54:1521–1526
Myhre S, Tveit H, Mollestad T, Laegreid A (2006) Additional gene ontology structure for improved biological reasoning. Bioinformatics 22:2020–2027
Olishevska S, Nickzad A, Déziel E (2019) Bacillus and Paenibacillus secreted polyketides and peptides involved in controlling human and plant pathogens. App Microbiol Biotech 103:1189–1215
Perkins DN, Pappin DJC, Creasy DM, Cottrell JS (1999) Probability based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis 20:3551–3567
Petersen TN, Brunak S, von Heijne G, Nielsen H (2011) SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat Methods 8:785–786
Pugsley AP (1993) The complete general secretory pathway in Gram-negative bacteria. Microbiol Rev 57:50–108
Qian CD, Wu XC, Teng Y, Zhao WP, Li O, Fang SG, Huang ZH, Gao HC (2012) Battacin (Octapeptin B5), a new cyclic lipopeptide antibiotic from Paenibacillus tianmuensis active against multidrug-resistant Gram-negative bacteria. Antimicrob Agents Chemother 56(3):1458–1465
Rivas FV, Tolia NH, Song JJ, Aragon JP, Liu J, Hannon GJ, Joshua-Tor L (2005) Purified Argonaute2 and an siRNA form recombinant human RISC. Nat Struct Mol Biol 12:340–349
Rybakova D, Cernava T, Köberl M, Liebminger S, Etemadi M, Berg G (2016) Endophytes-assisted biocontrol: novel insights in ecology and the mode of action of Paenibacillus. Plant Soil 405:125–140
Sáez-Nieto JA, Medina-Pascual MJ, Carrasco G, Garrido N, Fernandez-Torres MA, Villalón P, Valdezate S (2017) Paenibacillus spp. isolated from human and environmental samples in Spain: detection of 11 new species. New Microbes New Infect 19:19–27
Sanchez LA, Gomez FF, Delgado OD (2009) Cold-adapted microorganisms as a source of new antimicrobials. Extremophiles 13:111–120
Schneewind O, Missiakas DM (2012) Protein secretion and surface display in Gram-positive bacteria. Philos Trans R Soc Lond B Biol Sci 367:1123–1139
Selim S, Negrel J, Govaerts C, Gianinazzi S, Van Tuinen D (2005) Isolation and partial characterization of antagonistic peptides produced by Paenibacillus sp. strain B2 isolated from the sorghum mycorrhizosphere. App Environ Microbiol 71:6501–6507
Son SH, Khan Z, Kim SG, Kim YH (2010) Plant growth-promoting rhizobacteria, Paenibacillus polymyxa and Paenibacillus lentimorbus suppress disease complex caused by root-knot nematode and Fusarium wilt fungus. J Appl Microbiol 107:524–532
Takeuchi Y, Murai A, Takahara Y, Kainosho M (1979) The structure of permetin A, a new polypeptin type antibiotic produced by Bacillus circulans. J Antibiot 32:121–129
Tjalsma H et al (2004) Proteomics of protein secretion by Bacillus subtilis: separating the “secrets” of the secretome. Microbiol Mol Biol Rev 68:207–233
van Belkum MJ, Lohans CT, Vederas JC (2015) Draft genome sequences of Paenibacillus polymyxa NRRL B-30509 and Paenibacillus terrae NRRL B-30644, strains from a poultry environment that produce tridecaptin A and paenicidins. Genome Announc 3:e00372-15
Van Gerven N, Waksman G, Remaut H (2011) Pili and flagella biology, structure, and biotechnological applications. Prog Mol Biol Transl Sci 103:21–72
Vargas-Romero F, Guitierrez-Najera N, Mendoza-Hernández G, OrtegaBernal D, Hernández-Pando R, Castañón-Arreola M (2016) Secretome profile analysis of hypervirulent Mycobacterium tuberculosis CPT31 reveals increased production of EsxB and proteins involved in adaptation to intracellular lifestyle. Pathog Dis 74:ftv127
Xie JB, Zhang LH, Zhou YG, Liu HC, Chen SF (2012) Paenibacillus taohuashanense sp., nov. a nitrogen-fixing species isolated from rhizosphere soil of the root of Caragana kansuensis Pojark. Antonie Van Leeuwenhoek 102:735–741
Zainal Baharin NH, Khairil Mokhtar NF, Mohd Desa MN, Dzaraly ND, Muthanna AR, Al-Obaidi MM, Yuswan MH, Abbasiliasi S, Rahmad N, Wan Nur Ismah WA, Hashim AM, Mustafa S (2022) Inhibition mechanisms of secretome proteins from Paenibacillus polymyxa Kp10 and Lactococcus lactis Gh1 against methicillin-resistant Staphylococcus aureus and vancomycin-resistant Enterococcus. Asian Pac J Trop Biomed 12:483–494
Zengguo H, Duygu K, Liwen Z, Chunhua Y, Kari GC, Ahmed EY (2007) Isolation and identification of a Paenibacillus polymyxa strain that coproduces a novel lantibiotic and polymyxin. App Environ Microbiol 73:168–178
Zhang L, Ni H, Du X, Wang S, Ma XW, NüRnberger T, Guo HS, Hua C (2017) The Verticillium-specific protein VdSCP7 localizes to the plant nucleus and modulates immunity to fungal infections. New Phytol 215:368–381
Zijnge V, Kieselbach T, Oscarsson J (2012) Proteomics of protein secretion by Aggregatibacter actinomycetemcomitans. PLoS ONE 7:e41662
Zubair M, Muhamed SA, Khan FA, Zhao G, Menghwar H, Faisal M, Zhang H, Zhu X, Rasheed MA, Chen Y, Marawan MA, Chen H, Guo A (2020) Identification of 60 secreted proteins for Mycoplasma bovis with secretome assay. Microb Pathog 143:104135
Acknowledgements
The author acknowledges the Dept. of Bioengineering and Biotechnology, BIT Mesra for providing the infrastructure.
Funding
The study was funded by Department of Biotechnology, Government of India (grant number BT/RLF 2020–21).
Author information
Authors and Affiliations
Contributions
KK and PKS analyzed the secretome data. Y Ma edited the manuscript and helped in revision. RPS supervised the work and wrote the original draft.
Corresponding author
Ethics declarations
Ethics approval
This article does not contain any studies with human participants or animal performed by any of the authors.
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Kumari, K., Sharma, P.K., Ma, Y. et al. Secretome of Paenibacillus sp. S-12 provides an insight about its survival and possible pathogenicity. Folia Microbiol 68, 559–569 (2023). https://doi.org/10.1007/s12223-023-01032-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12223-023-01032-4