Abstract
Escherichia coli can cause serious infections in the neonates and pregnant women. Although E. coli is widely studied, E. coli lactose-negative (lac−) strains have been rarely described before. So, the aim of this study was to compare lac− and lactose-positive (lac+) E. coli strains in respect of antimicrobial susceptibility and the frequency of virulence genes (VGs). The study included 58 lac+ and 58 lac− E. coli strains isolated from pregnant women and neonates. Culture and the results of biochemical reactions were conducted for lac− and lac+ E. coli identification and differentiation. Disc diffusion test was performed to study the antimicrobial susceptibility of the isolates, and PCR was used to detect VGs. Resistance to at least one of the tested antibiotics was found among 14 (25.9%) E. coli lac+ and in 26 (44.9%) E. coli lac− strains. Both lac+ and lac− E. coli strains were mostly resistant to ampicillin (22.4 and 39.7%) and ticarcillin (20.7 and 39.7%). None of the tested strains produced extended-spectrum β-lactamases (ESBLs). Genes fimH, fimA, iutA, sfa/foc, neuC, ibeA, and hlyF were detected, respectively, in 96.6, 82.8, 32.8, 24.1, 22.4, 12.1, and 6.9% of lac+ E. coli strains and in 94.8, 86.2, 48.3, 19.0, 8.6, 8.6, and 1.7% of lac− strains. The antimicrobial susceptibility and the pathogenic potential of both tested groups of E. coli strains are similar. Therefore, omitting E. coli lac− strains as a potential etiological agent of infections may pose a threat to the health and life of both mothers and neonates.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Saprophytic Escherichia coli strains, colonizing the human alimentary tract, perform the function of commensals. In the case when those bacteria colonize the human organism beyond the alimentary tract, they become opportunistic pathogens (Tenaillon et al. 2010). Most often, E. coli strains can cause urinary tract infections (UTIs) and intestinal infections, both in children and in adults. In rarer cases, they are also responsible for generalized infections such as bacteremia, sepsis, and meningitis in neonates (Korczak et al. 2005; Obata-Yasuoka et al. 2002; Watt et al. 2003). An infection of a neonate may occur during the birth, when the child is exposed to a direct contact with bacteria present in the genital tract and sometimes also with microorganisms derived from the gastrointestinal tract of the woman giving birth (Watt et al. 2003). Colonization of the genital tract or anus of a pregnant woman by E. coli strains may also lead to infection of a neonate by ascending route. This infection happens most frequently as a result of premature rupture of membranes. Microorganisms from the lower part of the genital tract (vagina, uterine cervix) get to the amniotic sac and infect the fetus (Newton 2005). Also, UTIs occurring in pregnant women are serious threat to the fetus (Barber et al. 2013). Asymptomatic bacteriuria is the most often type of UTIs in pregnant women which, if not treated, in 20.0–30.0% of cases may lead to pyelonephritis. It may result in premature birth and developmental retardation or fetus necrosis, as well as a fetus with low birth weight (Janicka et al. 1997). Development of an infection largely depends on immunity of the host organism and the pathogenicity of the strain that causes it (Korczak et al. 2005). Adhesins, capsular antigen K1, hemolysins, aerobactin, and invasion protein IbeA play the essential role in the pathogenesis of infections caused by E. coli (Johnson 1991). Most E. coli rods are biochemically active. Their most important traits are the ability to decompose tryptophan into indole and to ferment lactose. However, in clinical material, the presence of non-reactive E. coli strains may be indicated. Their identification is difficult due to low metabolic activity, including the lack of the ability to lactose fermentation (Gadage et al. 2014). Lactose-negative (lac−) strains of E. coli, just as lactose-positive (lac+) strains, can have a number of virulence factors (VFs) and exhibit antibiotic resistance, as well as induce infections (Nicoletti et al. 1988). In the available references, there is little information about E. coli lac− strains. There are also no results of studies comparing these strains with E. coli lac+. Accordingly, the aim of this study was to investigate E. coli lac− strains isolated from pregnant women and neonates in respect of antibiotic susceptibility, the ability to produce the extended-spectrum β-lactamases (ESBLs), and the frequency of genes encoding selected VFs. The obtained data were compared with results obtained for E. coli lac+ strains.
Material and methods
Bacterial strains and its identification
The research material enclosed 116 genetically unrelated E. coli strains (58 lac+ and 58 lac−) isolated from pregnant women and neonates hospitalized at the Dr. Biziel University Hospital No.2 in Bydgoszcz. E. coli lac+ strains from pregnant women were obtained from rectal swabs (n = 26), vaginal swabs (n = 12), and urine (n = 8) and, in the case of neonates, from nasal cavities (n = 10) and urine (n = 2). In turn, E. coli lac− strains were isolated from rectal swabs (n = 52) and urine (n = 1) of pregnant women and from nasal cavities (n = 4) and urine (n = 1) of neonates.
E. coli species identification was based upon colony morphology, the ability to ferment lactose on the MacConkey agar (Becton Dickinson), and the results of biochemical reactions obtained in the VITEK®2 Compact (bioMérieux). E. coli lac+ and lac− strains were differentiated on the basis of colony color on the MacConkey agar and the result of the Taxo ONPG Disks test (Becton Dickinson).
Antimicrobial susceptibility testing and detection of ESBLs
The susceptibility of E. coli strains to selected antibiotics was assessed with the Kirby-Bauer disk-diffusion method on the Mueller-Hinton agar (Becton Dickinson) according to the recommendations of the European Committee on Antimicrobial Susceptibility Testing (EUCAST 2013). E. coli ATCC 25922 was used for quality-control purposes.
The production of ESBLs was assessed using the double-disk synergy test (Jarlier et al. 1988). E. coli ATCC 25922 (not producing ESBLs) and Klebsiella pneumoniae ATCC 700603 (producing ESBLs) were used as reference strains.
Detection of virulence genes
In order to detect VGs, isolation of DNA and multiplex PCR reactions were performed. DNA from the tested E. coli strains was isolated using the Genomic Mini kit (A&A Biotechnology). The presence of genes encoding the following VFs: type 1 fimbriae (fimA and fimH), S and F1C fimbriae (sfa/foc), antigen K1 (neuC), receptor for aerobactin (iutA), and protein IbeA (ibeA), hemolysin HlyF (hlyF), was detected with multiplex PCR, according to the authors’ methodology described before by Kaczmarek et al. (2012). The E. coli BEN2908 strain was used as positive control for the fimA, fimH, neuC, hlyF, iutA, and ibeA genes and strain 536 for sfa/foc. As a negative control, a reaction mixture without DNA was included in the experiment.
Statistical analysis
Statistical analysis of the results was carried out using the chi-square test and Fisher exact test, at the significance level α = 0.05. Calculations were made with the program Statistica 10 PL (StatSoft, Poland).
Results
Antibiotic susceptibility analysis
It was indicated that E. coli strains, both lac+ and lac−, were mostly resistant to ampicillin (22.4 and 39.7%) and ticarcillin (20.7 and 39.7%). Moreover, in the group of E. coli lac− strains, a high percentage of strains not susceptible to ticarcillin with clavulanic acid (19.0%) and cotrimoxazole (10.3%) was determined. From 1.7 to 6.9% of E. coli lac+ and lac− strains were resistant to the other tested antibiotics (Table 1). The statistically significant differences between E. coli lac+ and lac− were found in the resistance to ticarcillin (P < 0.05) and ticarcillin with clavulanic acid (P < 0.05). A significantly higher percentage of strains resistant to these antibiotics was recorded in the group of E. coli lac− strains.
None of the tested E. coli lac+ and lac− strains produced ESBL.
Among the tested E. coli strains, 20 antimicrobial resistance patterns were distinguished (Table 2). It was stated that 4 profiles (A–D) were common for both the tested groups of E. coli, whereas 7 (E–K) were typical only for E. coli lac+ strains and 9 (L–U) of E. coli lac−. Profile A occurred statistically significantly more frequently in E. coli lac+ (n = 43) than in E. coli lac− (n = 32) (P < 0.05) (Table 2). Among profiles typical only for E. coli lac+, two strains belonged to profile E and the other profiles (F–K) were represented by single E. coli strains. However, among profiles determined only for E. coli lac−, most isolates (n = 6) were classified to profile L, in which a statistically significant difference was indicated between E. coli lac+ and E. coli lac− strains (P < 0.05) (Table 2).
Moreover, 8 out of 20 drug-susceptibility profiles were represented also by E. coli strains intermediate to 1 (profiles E, G, I, O, P) and 2 antibiotics (profiles F, H, K). A higher percentage of antibiotic intermediate-susceptible strains was observed among E. coli lac+ strains than in the case of E. coli lac− strains (12.1 vs. 3.4%). One isolate of E. coli lac+ showed only an intermediate susceptibility to two tested antibiotics, at the same time not being resistant to any of the tested drugs (profile K) (Table 2).
Prevalence of VGs
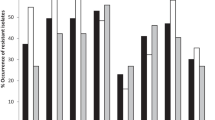
It was stated that the most common virulence genes (VGs), in E. coli lac+ and lac− strains, were fimH (96.6 and 94.8%, respectively) and fimA (82.8 and 86.2%, respectively) (Fig. 1). A considerable percentage of E. coli strains (32.8% lac+ and 48.3% lac−) had the gene iutA. On the other hand, the presence of genes sfa/foc, neuC, and ibeA was found, respectively, in 24.1, 22.4, and 12.1% of E. coli lac+ rods and in 19.0, 8.6, and 8.6% of lac−. The gene hlyF occurred the least commonly among the tested isolates (6.9% of lac+ and 1.7% of lac−). A statistically significant difference between the compared groups of E. coli lac+ and lac− strains was observed only in the case of the gene neuC (P < 0.05) (Fig. 1).
This study indicated that E. coli strains represented different profiles of genes encoding VFs. In the tested populations, 20 profiles of VGs were identified, including 9 common for E. coli lac+ and E. coli lac− strains as well as 8 typical only for E. coli lac+ and 3 of E. coli lac− (Table 3). Profile II occurred statistically significantly more frequently in E. coli lac− (n = 16) than in E. coli lac+ (n = 5) (P < 0.05) (Table 3).
For E. coli lac+ strains, a statistically insignificant (P > 0.05) but slightly positive correlation (r = 0.08) between the number of antibiotics to which the tested strains were resistant and the number of VGs which they had was showed (Fig. 2). From this, it follows that there was a very weakly drawn tendency to increase the virulence of strains along with an increase in their antibiotic resistance. In contrast, for E. coli lac− strains, a statistically insignificant (P > 0.05) but slightly negative correlation (r = −0.03) was shown (Fig. 3).
Discussion
E. coli should be ranked into the most important bacterial agents that can cause life-threatening infections in the perinatal period (Watt et al. 2003). These bacteria may be the cause of meningitis in neonates (neonatal meningitis-associated E. coli, NEMEC), bacteriemia and sepsis (sepsis-associated E. coli, SEPEC), and urinary tract infections (uropathogenic E. coli, UPEC) (Obata-Yasuoka et al. 2002; Watt et al. 2003; Alemu et al. 2012).
The analysis of the drug-susceptibility profiles of pathogenic bacterial strains enables the constant monitoring of their resistance to antibiotics. However, difficulty results from the fact that the authors most often do not report to which group (lac+ or lac−) they classified the tested E. coli strains. Therefore, when discussing the obtained present results, it is impossible to state to which population of E. coli we refer.
Susceptibility of E. coli strains to semi-synthetic penicillins is diverse in the world. In Poland (Janicka et al. 1997), Nigeria (Poey et al. 2012), and Ethiopia (Alemu et al. 2012; Tadesse et al. 2014), it was estimated that 80.8, 95.3, and 68.8–100% of E. coli strains were resistant to ampicillin, respectively. A lower percentage of E. coli strains resistant to ampicillin isolated from the vagina and rectum of pregnant women (39.9%), as well as those not pregnant (48.6%), was found by Hilbert et al. (2009) and Villar et al. (2013) and in the present study (31.0%). Moreover, in this study, a statistically significant difference was indicated between E. coli lac+ and lac− strains in resistance to ticarcillin (20.7 vs 39.7%). Also, a low percentage of E. coli strains resistant to penicillins with inhibitors of beta-lactamases and cephalosporins were found in this study. Significant differences between E. coli lac+ and lac− strains were found only in the case of resistance to ticarcillin with clavulanic acid (0.0 vs 19.0%). Similar results were also obtained in the authors’ earlier study (Kaczmarek et al. 2011), as well as in studies by other authors (Barcaite et al. 2012), irrespective of the fact if the strains were isolated from the rectum, vagina, or urine of pregnant women.
In the presented study, all the E. coli lac+ and lac− strains were susceptible to imipenem, amikacin, and tigecycline, which was also indicated in several other centers in Poland (Daniluk et al. 2008; Cisowska et al. 2001; Sobieszczańska et al. 2003; Sękowska et al. 2009). All the tested E. coli lac− strains and 94.8% of lac+ strains were also susceptible to aztreonam. Similar results were recorded in the earlier studies in Poland (Kaczmarek et al. 2011; Cisowska et al. 2001).
Moreover, a high percentage of E. coli strains from both tested groups susceptible to ciprofloxacin, norfloxacin, gentamicin, tobramycin, chloramphenicol, and trimethoprim/sulfamethoxazole was observed in the present study. An equally high percentage of strains susceptible to ciprofloxacin, with no resistance to gentamicin and amikacin, was isolated from neonates in Lithuania (98.3%) (Tamelienė et al. 2012). A high percentage of strains susceptible to ciproflofloxacin and aminoglycosides was also recorded in E. coli isolated from the vagina and rectum of pregnant women (Barcaite et al. 2012). Mehta et al. (2012) and Enayat et al. (2008) recorded a lower percentage of E. coli strains susceptible to fluoroquinolones and trimethoprim/sulfamethoxazole, respectively. Other authors that used E. coli isolated from the vagina of pregnant women reported almost 92% of the strains susceptible to cotrimoxazole (Spaetgens et al. 2002).
ESBLs producing E. coli strains are a serious therapeutic and epidemiological problem (Lina et al. 2014). In this study, none of the tested strains produced ESBLs. In the earlier study, also, no strains producing those enzymes were detected in E. coli isolated from pregnant women and neonates (Kaczmarek et al. 2011). In contrast, Villar et al. (2013) found the ability to produce ESBL enzymes in 5.4% of E. coli strains isolated from pregnant women.
The knowledge about etiological agents of an infection and their resistance to drugs, as well as the identification of traits responsible for the virulence of bacteria, is of the utmost importance in predicting the clinical course of an infection. The information about the virulence of the E. coli strain also facilitates making proper therapeutic decision.
In this study, in both groups of E. coli rods, more than 80% of strains having the gene fimA encoding the major structural subunit of type 1 fimbriae were identified. A similar result was obtained in the authors’ earlier study (Kaczmarek et al. 2012), as well as in the study by Micenková et al. (2014). On the other hand, the presence of the gene fimH determining the biosynthesis of the adhesin of those fimbriae was found in more than 90% of all the tested E. coli strains in the present study. Similarly, other authors indicated the presence of this gene among 90–100% of strains collected both from pregnant (Obata-Yasuoka et al. 2002; Poey et al. 2012; Al-Mayahie 2013) and not pregnant women (Obata-Yasuoka et al. 2002; Al-Mayahie 2013).
S- and F1C/S-related fimbriae are encoding by the genes sfa and foc, respectively (Juskova and Ciznar 1994). Watt et al. (2003) and Al-Mayahie (2013) indicated the presence of genes encoding S and F1C fimbriae in 48–60% of strains isolated from the vagina of pregnant women. In this study, the frequency of the gene sfa/foc was lower and amounted to 24.1% among E. coli lac+ strains and 19.0% among lac− strains.
In the presented study, the occurrence of the gene hlyF, encoding hemolysin F, was found in only 4.3% of strains, and it occurred more often in lac+ strains. In an earlier study, the percentage of strains that had the gene hlyF was significantly higher in E. coli K1 (28.4%), in comparison with the group of bacteria without this antigen (6.0%) (Kaczmarek et al. 2012).
E. coli producing the K1 antigen are responsible for about 80% of cases of meningitis in neonates (Watt et al. 2003). Moreover, they may be the cause of bacteremia, sepsis, and UTIs, both in children and adults (Alemu et al. 2012). The presence of the gene neuC, encoding the capsule K1, was higher in strains isolated from neonates with meningitidis (92%) (Ewers et al. 2007; Watt et al. 2003) than in strains isolated from the vagina, rectum, or urine of pregnant women (Watt et al. 2003). In the present study, E. coli lac+ strains had significantly more often gene neuC (22.4%) in comparison with the group of lac− strains (8.6%).
This study indicated the presence of the gene iutA, encoding the aerobactin receptor, in more than 30% of the tested E. coli lac+ strains and in almost 50% of E. coli lac− strains. A similar result was obtained by Poey et al. (2012) and Watt et al. (2003). In the study by Obata-Yasuoka et al. (2002), there were more often isolated strains having the gene-coding aerobactin from non-pregnant women than from pregnant ones.
Invasive protein IbeA takes part in the penetration of E. coli into brain microvascular endothelial cells. In the present study, the gene ibeA occurred only in 12.1% of E. coli lac+ strains and 8.6% of lac− strains, but none of the tested strains came from blood. In the earlier work, a significantly higher frequency of this gene was indicated among E. coli K1 (35.8%) than in strains without the capsule K1 (4.5%) (Kaczmarek et al. 2012).
E. coli rods may cause serious parenteral infections, and they are particularly dangerous for pregnant women and neonates. In the present study, it was indicated that there were more strains resistant to at least one antibiotic in the group of lac− isolates, and the frequency of genes encoding chosen virulence factors was similar in both groups of strains. E. coli lac+ and lac− strains had also common profiles of drug susceptibility and tested VGs. This may indicate their very similar pathogenic potential.
Conclusion
It is essential to pay attention to the presence of lac− strains of E. coli in clinical material in the course of diagnostic procedures and to carry out their species identification. Omitting E. coli lac− strains as a potential etiological agent of infections may pose a threat to the health and life of both mothers and newborn babies.
References
Al-Mayahie SM. 2013 Vaginal colonization by pap G allele II+ Escherichia coli isolates from pregnant and nonpregnant women as predisposing factor to pyelonephritis. Infect Dis Obstet Gynecol ; doi:10.1155/2013/860402
Alemu A, Moges F, Shiferaw Y, Tafess K, Kassu A, Anagaw B, Agegn A (2012) Bacterial profile and drug susceptibility pattern of urinary tract infection in pregnant women at University of Gondar Teaching Hospital, Northwest Ethiopia. BMC Res Notes 5:197. doi:10.1186/1756-0500-5-197
Barber AE, Norton JP, Spivak AM, Mulvey MA (2013) Urinary tract infections: current and emerging management strategies. Clin Infect Dis 57:719–724. doi:10.1093/cid/cit28
Barcaite E, Bartusevicius A, Tameliene R, Maleckiene L, Vitkauskiene A, Nadisauskiene R (2012) Group B Streptococcus and Escherichia coli colonization in pregnant women and neonates in Lithuania. Int J Gynaecol Obstet 117:69–73. doi:10.1016/j.ijgo.2011.11.016
Cisowska A, Ruczkowska J, Doroszkiewicz W (2001) Wrażliwość na antybiotyki szczepów Escherichia coli z otoczkowym antygenem K1 izolowanych z zakażeń układu moczowego. Med Dośw Mikrobiol 53:345–355
Daniluk T, Ściepuk M, Fiedoruk K, Zaremba ML, Rożkiewicz D, Ołdak E. Częstość występowania i wrażliwość na antybiotyki szczepów Escherichia coli i Klebsiella pneumoniae izolowanych od dzieci hospitalizowanych w Uniwersyteckim Szpitalu Dziecięcym w latach 2004–2007. Now Lek 2008; 77:265–72 (article in Polish). http://www.nowinylekarskie.ump.edu.pl/uploads/2008/4/265_4_77_2008.pdf Accessed 30 September 2015
Enayat K, Fariba F, Bahram N (2008) Asymptomatic bacteriuria among pregnant women referred to outpatient clinics in Sanandaj, Iran. Int Braz J Urol 34 (6):699–707
Ewers C, Li G, Wilking H, Kieβling S, Alt K, Antáo EM, Laturnus C, Diehl I, Glodde S, Homeier T, Böhnke U, Steinrück H, Philipp HC, Wieler LH (2007) Avian pathogenic, uropathogenic, and newborn meningitis-causing Escherichia coli: how closely related are they? Int J Med Microbiol 297:163–176. doi:10.1016/j.ijmm.2007.01.003
Gadage D, Wankhade A, Muley VV, Paralikar AV, Bhore AV (2014) Are inactive E. coli always commensals? Sch J App Med Sci 2:426–427 http://saspublisher.com/wp-content/uploads/2014/03/SJAMS-21D426-427.pdf Accessed 30 September 2015
Hilbert DW, Paulish TE, Mordechai E, Adelson ME, Gygax SE, Trama JP (2009) Antimicrobial non-susceptibility of cervico-vaginal and rectal Escherichia coli isolates is associated with phylogeny and plasmid carriage. Eur J Clin Microbiol Infect Dis 28:1399–1403. doi:10.1007/s10096-009-0788-3
Janicka G, Wojciechowska D, Hareńska K, Porada J, Kłyszejko C (1997) The resistance to betalactam antibiotics of lactose-positive and lactose-negative strains of Escherichia coli. Acta Microbiol Pol 46:399–403 (PMID:9516987)
Jarlier V, Nicolas M, Fourier G, Philippon A (1988) Extended broad-spectrum beta-lactamases conferring transferable resistance to newer beta-lactam agents in Enterobacteriaceae: hospital prevalence and susceptibility patterns. Rev Infect Dis 10:867–878 PMID:3263690
Johnson J (1991) Virulence factors in Escherichia coli urinary tract infection. Clin Microbiol Rev 4:80–128. doi:10.1128/CMR.4.1.80
Juskova E, Ciznar I (1994) Occurrence of genes for P and S fimbriae and hemolysin in urinary Escherichia coli. Folia Microbiol 39:159–161. doi:10.1007/BF02906814
Kaczmarek A, Budzyńska A, Gospodarek E (2011) Antimicrobial sensitivity of Escherichia coli straind with K1 antigen isolated from pregnant women and newborns. Med Dosw Mikrobiol 63:121–130 (article in Polish); http://www.medmikro.org/przeglad-epidemiologiczny-artykuly?year=2011&subissue=2 Accessed 30 September 2015
Kaczmarek A, Budzyńska A, Gospodarek E (2012) Prevalence of genes encoding virulence factors among Escherichia coli with K1 antigen and non-K1 E. coli strains. J Med Microbiol 61:1360–1365. doi:10.1099/jmm.0.044263-0
Korczak B, Frey J, Schrenzel J, Schrenzel J, Pluschke G, Pfister R, Ehricht R, Kuhnert P (2005) Use of diagnostic microarrays for determination of virulence gene patterns of Escherichia coli K1, a major cause of neonatal meningitis. J Clin Microbiol 43:1024–1031. doi:10.1128/JCM.43.3.1024-1031.2005
Lina TT, Khajanchi BK, Azmi IJ, Islam MA, Mahmood B, Akter M, Banik A, Alim R, Navarro A, Perez G, Cravioto A, Talukder KA et al (2014) Phenotypic and molecular characterization of extended-spectrum beta-lactamase-producing Escherichia coli in Bangladesh. PLoS One 9:e108735. doi:10.1371/journal.pone.0108735
Mehta M, Bhardwaj S, Sharma J (2012) Prevalence and antibiotic susceptibility pattern of multi-drug resistant Escherichia coli isolates from urinary tract infection (UTI) patients. Int J Life Sci Pharm Res 2:6–11 http://ijlpr.com/admin/php/uploads/131_pdf.pdf Accessed 30 September 2015
Micenková L, Štaudová B, Bosák J, Mikalová L, Littnerová S, Vrba M, Ševčíková A, Woznicová V, Šmajs D (2014) Bacteriocin-encoding genes and ExPEC virulence determinants are associated in human fecal Escherichia coli strains. BMC Microbiol 14:109. doi:10.1186/1471-2180-14-109
Newton ER (2005) Preterm labor, preterm premature rupture of membranes, and chorioamnionitis. Clin Perinatol 32:571–600. doi:10.1016/j.clp.2005.05.001
Nicoletti M, Superti F, Conti C, Calconi A, Zagaglia C (1988) Virulence factors of lactose-negative Escherichia coli strains isolated from children with diarrhea in Somalia. J Clin Microbiol 26:524–529 PMCID: PMC266325
Obata-Yasuoka M, Ba-Thein W, Tsukamoto T, Yoshikawa H, Hayashi H (2002) Vaginal Escherichia coli share common virulence factor profiles, serotypes and phylogeny with other extraintestinal E. coli. Microbiology 148:2745–2752. doi:10.1099/00221287-148-9-2745
Poey ME, Albini M, Saona G, Laviña M (2012) Virulence profiles in uropathogenic Escherichia coli isolated from pregnant women and children with urinary tract abnormalities. Microb Pathog 52:292–301. doi:10.1016/j.micpath.2012.02.006
Sękowska A, Ibsz-Fijałkowska A, Gołdyn K, Gospodarek E (2009) Wrażliwość na wybrane tetracykliny pałeczek z rodziny Enterobacteriaceae. Med Dośw Mikrobiol 61:321–326 (article in Polish); http://www.medmikro.org/wrazliwosc-na-wybrane-tetracykliny-paleczek-z-rodziny-enterobacteriaceae?lang=pl Accessed 30 September 2015
Sobieszczańska B, Kowalska-Krochmal B, Mowszet K, Pytrus T (2003) Wrażliwość na antybiotyki i chemioterapeutyki enteroagregacyjnych szczepów Escherichia coli (EAEC) izolowanych od dzieci z biegunką. Przegl Epidemiol 57:499–503 (article in Polish); http://www.przeglepidemiol.pzh.gov.pl/wrazliwosc-na-antybiotyki-i-chemioterapeutyki-enteroagregacyjnych-szczepow-escherichia-coli-eaec-izolowanych-od-dzieci-z-biegunka?lang=pl Accessed 30 September 2015
Spaetgens R, DeBella K, Ma D, Robertson S, Mucenski M, Davies HD (2002) Perinatal antibiotic usage and changes in colonization and resistance rates of group B Streptococcus and other pathogens. Obstet Gynecol 100:525–533. doi:10.1016/S0029-7844(02)02068-9
Tadesse E, Teshome M, Merid Y, Kibret B, Shimelis T (2014) Asymptomatic urinary tract infection among pregnant women attending the antenatal clinic of Hawassa Referral Hospital. Southern Ethiopia BMC Res Notes 17:155. doi:10.1186/1756-0500-7-155
Tamelienė R, Barčaitė E, Stonienė D, Buinauskienė J, Markūnienė E, Kudrevičienė A, Vitkauskienė A, Jomantienė D, Nadišauskienė R (2012) Escherichia coli colonization in neonates: prevalence, perinatal transmission, antimicrobial susceptibility, and risk factors. Medicina (Kaunas) 48:71–76 http://medicina.lsmuni.lt/med/1202/12040501.pdf Accessed 30Sepember 2015
Tenaillon O, Skurnik D, Picard B, Denamur E (2010) The population genetics of commensal Escherichia coli. Nat Rev Microbiol 8:207–217. doi:10.1038/nrmicro2298
The European Committee on Antimicrobial Susceptibility Testing. Breakpoint tables for interpretation of MICs and zone diameters. Version 3.1, 2013. http://www.eucast.org. Accessed 30September 2015
Villar HE, Aubert V, Baserni MN, Jugo MB (2013) Maternal carriage of extended-spectrum beta-lactamase-producing Escherichia coli isolates in Argentina. J Chemother 25:324–327. doi:10.1179/1973947813Y.0000000081
Watt S, Lanotte P, Mereghetti L, Moulin-Schouleur M, Picard B, Quentin R (2003) Escherichia coli strains from pregnant women and neonates: intraspecies genetic distribution and prevalence of virulence factors. J Clin Microbiol 41:1929–1935. doi:10.1128/JCM.41.5.1929-1935.2003
Acknowledgements
We are grateful to Dr. P. Germon (Unité Infectiologie Animale et Santé Publique, France) for the E. coli BEN2908 strain and Dr. U. Dobrindt (Institut für Molekulare Infectionsbiologie, Zentrum für Infektionsforschung, Universität Würzburg, Germany) for the E. coli 536 strain. We are also grateful to Prof. W. Szymanski (Department of Obstetrics and Gynecology, Ludwik Rydygier Collegium Medicum in Bydgoszcz, Nicolaus Copernicus University in Torun, Poland), Dr. P. Korbal (Department of Neonatology), M.Sc. J. Kochanowska (Department of Microbiology), and the other staff at the Dr. Jan. Biziel University Hospital No.2 in Bydgoszcz, Poland) for their clinical bacterial samples. Without them, we would not have been able to obtain these collections.
This research was financially supported by the Nicolaus Copernicus University with funds from the maintenance of the research potential of the Department of Microbiology.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethics statement
This article does not contain any studies with human participants or animals performed by any of the authors.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Kaczmarek, A., Skowron, K., Budzyńska, A. et al. Virulence genes and antimicrobial susceptibility of lactose-negative and lactose-positive strains of Escherichia coli isolated from pregnant women and neonates. Folia Microbiol 62, 363–371 (2017). https://doi.org/10.1007/s12223-017-0506-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12223-017-0506-y