Abstract
We used SDS-polyacrylamide gel electrophoresis to investigate the outer membrane proteins (OMPs) band composition of 19 Escherichia coli K1 strains that have spontaneously lost the ability to form K1 polysaccharide capsule (E. coli K1−) and demonstrated different degrees of susceptibility to the bactericidal action of normal human serum. Presented results showed that there were differences between E. coli K1− strains in OMPs expressing capacity. The analysis performed on OMPs has not revealed a direct association between the different OMPs band composition and the susceptibility of these strains to the serum.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Uropathogenic Escherichia coli strains are the major cause of urinary tract infections (UTIs) and are responsible for approximately 70–95 % of community acquired and 50 % of nosocomial UTIs (Kucheria et al. 2005). Mostly, these infections arise in the bladder (cystitis); however, in cases where the kidneys (pyelonephritis) are involved, the course of the disease is usually more severe. Frequent causative agent of childhood pyelonephritis is K1 capsular polysaccharide expressing E. coli rod (Johnson 1991). Moreover, E. coli K1 strains are a particularly important cause of neonatal meningitis (more than 80 % of the cases) and are commonly associated with neonatal septicaemia. Typically, these pathogens are capable of invading the brain of the newborns as a complication of the bloodstream dissemination following the colonization of the gastrointestinal tract (Nassif et al. 2002; Wooster et al. 2006; Parthasarathy et al. 2007). In order to invade the meninges, the bacteria must multiply in the blood and reach a high degree of bacteraemia (>104 colony forming units (CFU)/mL of blood) and, after this, cross the blood–brain barrier. Pathogens presenting brain tropism must avoid the host’s innate defence mechanisms, such as the complement (C) system and opsonophagocytosis, to survive in the bloodstream. This suggests that these microorganisms possess very specific virulence factors which are essential for meningeal invasion.
One of the significant factors in the pathogenicity of E. coli K1 strains is their resistance to the action of the C. The mechanism of the resistance to the bactericidal effect of the serum is not completely explained but it is known that this phenomenon has a multifactor basis. The organisation of the bacterial outer membrane and the structures such as bacterial capsule, outer membrane proteins (OMPs), and O-specific side chains of lipopolysaccharides (LPS) are the factors determining the susceptibility of the bacteria to the bactericidal action of the C (Taylor 1992, 1995).
The capsular polysaccharide produced by E. coli K1 strains is a homopolymer of α-2,8-linked sialic acid (N-acetylneuraminic acid, Neu5Ac, NeuAc) residues and is structurally and immunologically identical to the capsular polymer of Neisseria meningitidis group B, Pasteurella haemolytica A2, and Moraxella nonliquefaciens (Ferrero and Aparicio 2010). This polysaccharide can mimic the mammalian polysialic acid structure that can be found predominantly on the neural cell adhesion molecule (NCAM), which is responsible for mediating several neuronal functions by controlling intercellular adhesion, neurite outgrowth, cell migration, proliferation, and survival. Due to biochemical similarity with polysialylated form of humans NCAM, bacterial α-2,8-linked sialic acid is poorly immunogenic in humans and, hence, its immunological tolerance is observed.
The presence of NeuAc on the surface of the bacteria impedes the action of the C by enhancing the binding of factor H to C3b on the outside of the cell and, consequently, by reducing the amplification of C3b production and downregulation of the activity of the alternative pathway (Rautemaa and Meri 1999; Kugelberg et al. 2008). The protection against serum killing is also the result of the interaction between C4b-binding protein (C4bp), the predominant serum inhibitor of C3b activation via the classical pathway, and the outer membrane protein A (OmpA) of E. coli K1 rods (Wooster et al. 2006; Prasadarao et al. 2002). Moreover, the component of the bacterial capsule, α-2,8-linked Neu5Ac, can provide resistance against the C attack by inhibiting the insertion of membrane attack complex (MAC) into the bacterial membrane. Therefore, an increased synthesis of K1 capsule has a significant impact on the activation of the C, opsonization, and virulence of E. coli K1 rods. On the other hand, E. coli K1 strains demonstrate different degrees of sensitivity to the bactericidal action of the serum, which indicates that other outer membrane structures may also play a crucial role in the determination of bacterial resistance to the serum (Cisowska et al. 2004; Bugla-Płoskońska et al. 2006). Vermeulen et al. (1988) reported that there is a threshold amount of K1 capsular polysaccharides required for the protection from lysis by the serum. In addition, these authors showed that strains generating the same quantity of K1 polysaccharide but possessing a rough or a smooth LPS phenotype varied in the susceptibility to serum killing; specifically, the strain that lacked O side chain of LPS was more sensitive to serum bactericidal killing. Our previous results concerning Salmonella O48 indicate that neither the presence of NeuAc in LPS nor the length of the O-specific part of LPS containing NeuAc play a decisive role in determining bacterial resistance to the bactericidal activity of C and that the presence of NeuAc in the structure of LPS is not sufficient to block the activation of the alternative pathway of C (Bugla-Płoskońska et al. 2010). The mechanism of bacterial resistance to the bactericidal effect of the serum is still not fully understood and from the above results it may be concluded that other components of the bacterial cell envelope, for example OMP, could probably influence the C systems. Weiser and Gotschlich (1991) demonstrated that OmpA increased E. coli K1 rods resistance to the bacteriolytic activity of the C. They observed that the OmpA mutant with insertion in the ompA gene that completely eliminated the expression of OmpA was more sensitive to the bactericidal effect of human serum in regards to the classical pathway of C activation and suggested that OmpA may serve to stabilise the outer membrane, making it more resistant to the effects of C or may bind antibodies that block serum killing.
Our previous results demonstrated the diverse susceptibility of E. coli K1 strains to the bactericidal action of normal human serum (NHS) and showed the role of the particular mechanisms of C activation in the process of NHS-mediated killing of these rods (Cisowska et al. 2004; Bugla-Płoskońska et al. 2006). They confirmed that the presence of the K1 polysaccharide capsule does not play a decisive role in the determination of bacterial resistance to the lytic action of the serum. Subsequently, we have obtained K1− mutant forms (E. coli K1−) that have spontaneously lost the ability to produce K1 capsular antigen. In the present study, we have used sodium dodecyl sulphate-polyacrylamide gel electrophoresis (SDS-PAGE) in order to investigate outer OMPs band composition of E. coli K1− strains and to discern the relationship between OMPs pattern and susceptibility to NHS.
Material and methods
Bacterial strains
Eighteen E. coli K1 strains isolated from urine of children with urinary tract infections and a standard E. coli B2095 K1 strain were used to obtain unencapsulated E. coli K1−. K1− mutant forms were derived by selection for resistance to the K1-specific bacteriophage.
Detection of capsular antigen K1 in E. coli
The K1-specific bacteriophage (K1 A) was used for the detection of capsular antigen K1 in accordance with the procedure used by Devine et al. (1990). The E. coli strains were grown overnight on nutrient agar, and then bacterial cells were inoculated into 4 mL of nutrient broth and incubated at 37 °C for 4 h. Afterwards, a single streak of each culture was applied to nutrient agar and allowed to dry. Once the streak was dry, 10 μL of bacteriophage suspension (8 × 1010 PFU/mL) was placed in the centre of the streak. After incubation at 37 °C for 18 h, the presence of K1 was indicated by a clear zone of bacterial lysis at the site of the placement of bacteriophage suspension. E. coli K1− forms were obtained from colonies growing in an otherwise clear zone of bacterial lysis where the K1 A specific bacteriophage was used. Once isolated, these colonies were tested three times to check whether the loss of the ability to produce K1 capsule is a permanent feature. The E. coli K1− colonies were inoculated into nutrient broth and incubated at 37 °C for 4 h; following this, a single streak of each culture was applied to nutrient agar and allowed to dry. Next, the phage K1 A suspension was placed in the centre of the streak. After incubation at 37 °C for 18 h, no zone of bacterial lysis was observed.
Phage K1 A and standard strain E. coli B2095 (O2:K1) used for the proliferation of the phage were obtained from the collection of Dr. G. Schmidt (Forschungsinstitut Borstel, Institut für Experimentealle Biologie und Medizin, Germany). The bacteriophage suspension was stored in 2-mL portions at 4 °C.
Serum
NHS was obtained from healthy adult volunteers untreated with any antimicrobial drugs. The samples of NHS were collected, pooled, and stored in 0.5 mL portions at −70 °C. A suitable volume of the serum was thawed immediately before experiments. Each portion was used only once.
Bactericidal activity of serum
The bactericidal activity of NHS was determined as described previously (Cisowska et al. 2004). Briefly, the strains were grown overnight and then bacterial cells of early exponential growth phase were transferred to fresh nutrient broth and incubated at 37 °C for 0.5 h. After incubation, the bacterial cells were centrifuged (4,000 rpm for 20 min). Next, 0.5 mL of bacterial suspension was added to the dividing NHS (serum was diluted with 0.1 mol/L NaCl) giving the final serum concentration of 50 %; the samples were immediately collected and diluted and then cultured on nutrient agar plates for 18 h at 37 °C (0 min). Bacteria with NHS were incubated in a water bath at 37 °C; the samples were collected after 30, 60, and 180 min and then they were diluted and cultured in the same manner as described above. The number of CFU at 0 min was assumed as 100 %. Strains with a survival ratio of not less than 50 % after 180 min of incubation in the sera were regarded as resistant.
Isolation of the OMPs
The OMPs were isolated using procedure described by Murphy and Bartos (1989) with our own modifications. For the OMP isolation, we used bacteria grown in 100 mL brain heart infusion broth (Difco) at 37 °C for 18 h. After incubation, the bacterial cells were centrifuged (4,000 rpm at 4 °C for 20 min) and the resulting pellet was suspended in 2.5 mL of buffer B (1 mol/L sodium acetate, 0.001 mol/L β-mercaptoethanol; pH 4.0). Afterwards, 22.5 mL of the solution containing 5 % Zwittergent Z 3-14® (Calbiochem–Behring) and 0.5 mol/L CaCl2 was added. This solution was stirred at room temperature for 1 h. To precipitate nucleic acids, a volume of 6.25 mL of 96 % cold ethanol was added very slowly. Subsequently, the mixture was centrifuged at 12,300 rpm at 4 °C for 10 min. The remaining proteins were precipitated by the addition of 93.5 mL of 96 % cold ethanol and by centrifugation at 12,300 rpm at 4 °C for 20 min. The pellet was suspended in 5 mL of buffer Z (0.05 % Zwittergent, 0.05 mol/L Tris and 0.01 mol/L EDTA; pH 8.0) and stirred for 1 h at room temperature. The solution was kept at 4 °C overnight and centrifuged at 8,700 rpm at 4 °C for 10 min to obtain OMPs in the supernatant. Following the isolation of the OMPs, we checked the enzymatic activity of succinic dehydrogenase, a marker of cytoplasmic protein membranes, in the soluble fraction of buffer Z using the method of Rockwood et al. (1987). The Zwittergent-extracted OMPs contained no detectable succinic dehydrogenase activity, indicating that these preparations were not contaminated by cytoplasmic membrane.
Protein quantification was performed with the bicinchoninic acid (BCA) Protein Assay Kit (PIERCE®) with bovine serum albumine (Sigma) as the standard. The Pierce BCA Protein Assay is a detergent-compatible formulation based on BCA for the colorimetric detection and quantification of total protein concentration (PIERCE Instruction). The purple-coloured reaction product (reduction of Cu2+ to Cu1+ by protein in an alkaline medium) exhibits a strong absorbance at 562 nm with increasing protein concentration over a broad range of 20–20,000 μg/mL. OMPs after preparation in the sample buffer were loaded into each of the wells of the SDS-PAGE gels in comparable concentrations of 9–10 μg per well.
Polyacrylamide gel electrophoresis
SDS-PAGE was carried out on slabs with 12.5 % acrylamide as described by Laemmli (1970). Samples were applied to slabs after heating at 100 °C for 4 min. The Wide Molecular Weight Range Sigma Marker M4038 protein standard (6-205 kDa) was used for molar mass calibration. Electrophoresis was performed at 100 V for 25 min and then at 200 V for 45 min. After the electrophoresis, gel was stained for 1 h with a solution containing 25 % methanol, 10 % acetic acid, and 0.05 % Commasie Brilliant blue and decolorized with 10 % acetic acid for about 3 h (we used identical procedure for E. coli K1+; Cisowska et al. 2005). For the molecular analysis of OMPs BIO-CAPT v. 99 computer program as well as the BIO-1D++ v. 99 program (Vilber Lourmat, France; BioRad) were used.
Results
The results concerning the susceptibility to 50 % NHS of E. coli strains without K1 polysaccharide capsule (K1− forms) are enclosed in Table 1. The detailed results of susceptibility of E. coli K1+ strains to 50 % NHS were previously published (Cisowska et al. 2004); therefore, in Table 1 we presented only the survival percent after 3 h of incubation in NHS for K1+ strains in order to make the comparison between K1+ and K1− forms easier. The tested E. coli K1+ rods showed diverse susceptibility to the bactericidal action of NHS. Among 19 E. coli K1+ strains, eight (E. coli 988, 601, 529, 544, 1000, 315, 105, and 380) were sensitive to NHS. The remaining strains were resistant to NHS and the survival ratio of these strains after 180 min of incubation in serum ranged from 82.8 to 4,250.0 % of the initial cell number. Most of E. coli K1− isolates were sensitive to the bactericidal action of NHS (E. coli 380, 763, 848, 500, 5915, 309, 988, 601, 529, 158, 481, 544, 1000, 328, 315, 34, and 105). Moreover, in the case of some of them, NHS decreased the percent of survival to <0.0001 % even after 30–60 min of incubation. Two E. coli K1− strains were resistant to the serum and proliferated in NHS very intensively (E. coli 111 and B2095).
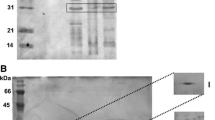
To investigate OMPs band patterns of E. coli K1− strains and evaluate whether there is a relationship between OMPs composition and susceptibility of these unencapsulated forms to bactericidal action of NHS, the SDS-PAGE was used. The electrophoretic band patterns for the OMPs of these rods and the molar mass of these proteins are shown in Figs. 1, 2, and 3.
The analysis performed on OMPs has not revealed a direct correlation between the various OMPs band patterns and the susceptibility of these strains to the serum. E. coli K1− strains sensitive to the lytic activity of NHS possessed between 12 and 22 different OMPs bands and the molar mass of these proteins ranged from 13.9 to 106.2 kDa (Figs. 1, 2, and 3). The greatest diversity in the quantity of possessed OMPs was observed for E. coli 763 K1− strain, which had 22 bands corresponding to proteins with molar mass ranging from 14.0 to 93.1 kDa (Fig. 3, lane 3). The lowest diversity was found for E. coli 1000 K1− strain. In this case, only 12 bands, corresponding to proteins with molar mass between 17.5 and 59.9 kDa, were obtained (Fig. 2, lane 3). Additionally, NHS-resistant K1− strains E. coli B2095 and 111 did not differ significantly from NHS-sensitive strains. It was noted also that E. coli K1− strains resistant to serum contained 13 (E. coli B2095; Fig. 2, lane 9) or 15 (E. coli 111; Fig. 1, lane 6) different OMPs with the molar mass ranging from 16.9 to 60.7 and 17.5 to 66.4 kDa, respectively. However, proteins with higher molar mass were detected only in certain K1− strains sensitive to the serum. These proteins were observed for example in the case of strains: E. coli 988 K1− (Fig. 1, lane 2; OMP: 102 and 75 kDa), E. coli 601 K1− (Fig. 1, lane 3; OMP: 102, 93, and 79 kDa), E. coli 529 K1− (Fig. 1, lane 4; OMP: 89 and 79 kDa), E. coli 315 K1− (Fig. 2, lane 5; OMP: 83 kDa), E. coli 34 K1− (Fig. 2, lane 6; OMP: 106, 97, 84, and 74 kDa), E. coli 763 K1− (Fig. 3, lane 3; OMP: 93, 86, and 77 kDa), E. coli 848 K1− (Fig. 3, lane 4; OMP: 105 and 78 kDa), or E. coli 309 K1− (Fig. 3, lane 7; 95 kDa).
The comparison of the outer membrane proteins patterns of the two serum-resistant K1− strains, E. coli B2095 (Fig. 2, lane 9) and 111 (Fig. 1, lane 6), shows a close correspondence. These rods mostly produced the same OMPs (60, 47, 43, 33, 31, and 22 kDa) and differed only in respect to a few of them (E. coli 111 – 66, 40, and 30 kDa proteins; E. coli B2095, 26 kDa protein). Since the bands corresponding to proteins with such molar masses were also detected in the group of NHS-sensitive K1− strains, we could not relate the occurrence of a specific OMP to serum susceptibility.
Several differences can be seen when SDS-PAGE patterns of the outer membrane proteins of both NHS-resistant K1+ and K1− E. coli B2095 forms are examined (Fig. 2, lanes 8 and 9). The number of OMPs generated by K1− mutant was comparable to that of the K1+ strain, but differed qualitatively. OMPs with the molar masses: 113, 93, 71, 50, 46, and 38 kDa were not produced by E. coli B2095 K1− form, and, additionally, some proteins (52, 47, 26, 23, and 16 kDa) were not formed by K1+ rods. Moreover, E. coli K1− mutant over-expressed some of the proteins (22, 33, and 35 kDa) as compared to K1+ form.
Discussion
During the immune responses, the ability to avoid killing by the serum C is an important evasion strategy of bacterial pathogens. The mechanism of the bacterial resistance to the bactericidal effect of the serum is not fully understood; however, it is known that the OMPs are one of the factors determining the resistance or sensitivity of bacteria to the bactericidal action of the C (Taylor 1992, 1995).
In the bloodstream, N. meningitidis is exposed to the antibody/complement-mediated lysis, as well as to opsonophagocytosis, and it seems that the quantity of the polysaccharide capsule that is produced influences the resistance of this pathogen to killing by the host (Uria et al. 2008). Only the encapsulated meningococci appear to survive in the blood and are able to interact with some negative regulators of the C cascade (factor H, C4bp), potentially leading to their increased survival (Hill et al. 2010). Factor H-binding protein, expressed by all meningococcal strains, can recruit C factor H, a key regulator of the alternative C pathway, which promotes serum resistance of these pathogens (Madico et al. 2006). Also, the Opc protein, another OMP commonly expressed by N. meningitidis strains, is able to bind vitronectin (Vn), which inhibits C5b-7 complex formation and C9 polymerization (Singh et al. 2010). As a consequence, Vn deposited on the bacterial surface inhibits MAC formation and protects the bacteria from MAC-mediated lysis. Furthermore, the major OMP of meningococci Por A can bind to the C regulator C4-binding protein and influence serum resistance; although, it may be inhibited by the capsule (Jarva et al. 2005). It must be mentioned, however, that Drogari-Apiranthitou et al. (2002) did not find a significantly reduced C3b deposition in two PorA(−) mutant strains and showed similar amounts of MAC deposition in PorA(+) and PorA(−) variants, regardless of the variance in their serum sensitivity.
In the present work, the OMPs of 19 E. coli K1− mutant strains were isolated and subjected to SDS-PAGE in order to characterise OMPs. In our opinion, the enhancement of the resistance to C-mediated bactericidal activity of the serum may be associated with the presence of the specific OMPs in bacterial outer membrane. It has been demonstrated that OmpA increases the resistance to bactericidal effect of the serum by the activation of the C classical pathway and is the source of E. coli K1 virulence (Weiser and Gotschlich 1991). Relationship between the resistance to the C, virulence of E. coli strains, and OMPs banding pattern was also noted by Chaffer et al. (1999). These authors observed that highly resistant to bactericidal effect of the C and highly virulent E. coli rods had a weak peptide band located at 35 kDa, while the occurrence of a strong 35 kDa OMP band was associated with a lower virulence and C susceptibility. In our previous work, we noticed the presence of the peptide band located at 35 kDa both in E. coli strains that were resistant and sensitive to the normal bovine serum and normal cord serum (Cisowska et al. 2005). Also, in the present study both of the NHS-resistant K1− mutant strains, E. coli B2095 and 111, and NHS-sensitive K1− strains (158, 309, 481, 848, 5915, 105, 315, 328, 380, 544, 601, 988, and 1000) possessed peptide band located at ∼35 kDa. Only four K1− strains (34, 500, 763, and 529) did not have ∼35 kDa OMP, while their K1+ forms generated a protein band with such molar mass (Cisowska et al. 2005).
Other investigators associated the resistance of E. coli rods to the C-mediated serum killing with 25 kDa OMP (Moll et al. 1980). In the case of the tested K1− mutant strains, a peptide band located at ∼25 kDa was detected only in a few NHS-sensitive E. coli (158, 380, 544, 988, 1000, 34, 5915, and 848). However, both of the serum-resistant K1− strains, E. coli B2095 and 111, did not produce this protein. Moreover, in our previous work, we noticed the presence of the peptide band located at ∼25 kDa in E. coli K1+ strains both resistant (111, 481, 763, 34, and 5915) and sensitive (328, 529, 380, 544, 988, and 1000) to bactericidal effect of the serum (Cisowska et al. 2005).
The comparison of the OMPs patterns of E. coli K1+ (Cisowska et al. 2005) and their K1− mutant forms, both the NHS-resistant and NHS-sensitive strains, showed that they differed in the amount and the type of produced OMPs. Regardless of the susceptibility of E. coli K1+ rods to bactericidal activity of NHS, the majority of K1− mutants produced a smaller amount of OMPs. In particular, this difference can be seen in the case of E. coli 1000, 544, 328, and 158 K1− rods that produced even 23–26 OMPs less than the corresponding K1+ variants. In contrast, in only three unencapsulated K1− forms the number of distinct electrophoretic components increased. Serum-resistant E. coli 34 K1+ and 763 K1+ as well as NHS-sensitive E. coli 601 K1+ generated 16, 17, and 15 OMPs, respectively, while their K1− forms produced 21, 22, and 19 OMPs, correspondingly.
In conclusion, in our study, we have not found any similarities between OMPs’ band patterns of analysed group of E. coli K1− rods. Our study did not find a OMPs band pattern characteristic for the analysed group of E. coli K1− rods. The diversity of OMPs may indicate a significant role that these proteins play in bacterial resistance to the bacteriolytic action of the C. On the other hand, presumably, the proteins that sensitise the bacterial cell to the bactericidal action of the C can be located among the OMPs that were analysed in this study.
Abbreviations
- C:
-
Complement
- NHS:
-
Normal human serum
- OMP:
-
Outer membrane protein
- UPEC:
-
Uropathogenic Escherichia coli strains
- UTIs:
-
Urinary tract infections
References
Bugla-Płoskońska G, Cisowska A, Karpińska K, Jankowski S, Doroszkiewicz W (2006) The mechanisms of the activation of normal human serum complement by Escherichia coli strains with K1 surface antigen. Folia Microbiol 51:627–632
Bugla-Płoskońska G, Rybka J, Futoma-Kołoch B, Cisowska A, Gamian A, Doroszkiewicz W (2010) Sialic acid-containing lipopolysaccharides of Salmonella O48 strains—potential role in camouflage and susceptibility to the bactericidal action of normal human serum. Microb Ecol 59:601–613
Chaffer M, Heller ED, Schwartsburd B (1999) Relationship between resistance to complement, virulence and outer membrane protein patterns in pathogenic Escherichia coli O2 isolates. Vet Microbiol 64:323–332
Cisowska A, Bugla-Płoskońska G, Tichaczek-Goska D, Doroszkiewicz W, Jankowski S (2004) The susceptibility of Escherichia coli strains with sialic acid-containing lipopolysaccharides or capsules to the bactericidal action of normal human serum. 7th Conference on Molecular Biology in Diagnostics of Infectious Diseases and Biotechnology. SGGW, Warsaw, pp 41–47
Cisowska A, Bugla-Płoskońska G, Gamian A, Doroszkiewicz W, Jankowski S (2005) Relationship between susceptibility to bactericidal action of serum and outer membrane protein patterns in E. coli K1 strains. Pol J Environ Stud 14:476–482
Devine DA, Roberts AP, Rowe B (1990) Simple technique for detecting K1 antigen of Escherichia coli. J Clin Pathol 43:76–78
Drogari-Apiranthitou M, Kuijper EJ, Dekker N, Dankert J (2002) Complement activation and formation of the membrane attack complex on serogroup B Neisseria meningitidis in the presence or absence of serum bactericidal activity. Infect Immun 70:3752–3758
Ferrero MA, Aparicio LR (2010) Biosynthesis and production of polysialic acids in bacteria. Appl Microbiol Biotechnol 86:1621–1635
Hill DJ, Griffiths NJ, Borodina E, Virji M (2010) Cellular and molecular biology of Neisseria meningitidis colonization and invasive disease. Clin Sci 118:547–564
Jarva H, Ram S, Vogel U, Blom AM, Meri S (2005) Binding of the complement inhibitor C4bp to serogroup B Neisseria meningitidis. J Immunol 174:6299–6307
Johnson JR (1991) Virulence factors in Escherichia coli urinary tract infection. Clin Microbiol Rev 4:80–128
Kucheria R, Dasgupta P, Sacks SH, Khan MS, Sheerin NS (2005) Urinary tract infections: new insights into a common problem. Postgrad Med J 81:83–86
Kugelberg E, Gollan B, Tang CM (2008) Mechanisms in Neisseria meningitides for resistance against complement-mediated killing. Vaccine 26:134–139
Laemmli UK (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680–685
Madico G, Welsch JA, Lewis LA, McNaughton A, Perlman DH, Costello CE, Ngampasutadol J, Vogel U, Granoff DM, Ram S (2006) The meningococcal vaccine candidate GNA1870 binds the complement regulatory protein factor H and enhances serum resistance. J Immunol 177:501–510
Moll A, Manning PA, Timmis KN (1980) Plasmid-determined resistance to serum bactericidal activity: a major outer-membrane protein, the traT gene product, is responsible for plasmid-specified serum resistance in Escherichia coli. Infect Immun 28:359–367
Murphy T, Bartos LC (1989) Surface-exposed and antigenically conserved determinants of outer membrane proteins of Branhamella catarralis. Infect Immun 57:2938–2941
Nassif X, Bourdouloud S, Eugene E, Couraud PO (2002) How do extracellular pathogens cross the blood–brain barrier? Trends Microbiol 10:227–232
Parthasarathy G, Yao Y, Kim KS (2007) Flagella promote Escherichia coli K1 association with and invasion of human brain microvascular endothelial cells. Infect Immun 75:2937–2945
Pierce Instructions. Pierce®BCA Protein Assay kit. Thermo Scientific, Pierce Biotechnology.
Prasadarao NV, Blom AM, Villoutreix BO, Linsangan LC (2002) A novel interaction of outer membrane protein A with C4b binding protein mediates serum resistance of Escherichia coli K1. J Immunol 169:6352–6360
Rautemaa R, Meri S (1999) Complement-resistance mechanisms of bacteria. Microbes Infect 1:785–794
Rockwood D, Wilson MT, Darley-Usmar VM (1987) Isolation and characteristic of intact mitochondria. In: Darley-Usmar VM, Rickwood D, Wilson MT (eds) Mitochondria: a practical approach. IRL, Oxford, pp 1–16
Singh B, Y-Ch S, Riesbeck K (2010) Vitronectin in bacterial pathogenesis: a host protein used in complement escape and cellular invasion. Mol Microbiol 78:545–560
Taylor PW (1992) Complement-mediated killing of susceptible Gram-negative bacteria: an elusive mechanism. Exp Clin Immunogenet 9:48–56
Taylor PW (1995) Resistance of bacteria to complement. In: Roth JA (ed) Virulence mechanism of bacterial pathogen. Am Soc Microbiol Washington DC.
Uria MJ, Zhang Q, Li Y, Chan A, Exley RM, Gollan B, Chan H, Feavers I, Yarwood A, Abad R, Borrow R, Fleck RA, Mulloy B, Vazquez JA, Tang CM (2008) A generic mechanism in Neisseria meningitidis for enhanced resistance against bactericidal antibodies. J Exp Med 205:1423–1434
Vermeulen C, Cross A, Byrne WR, Zollinger W (1988) Quantitative relationship between capsular content and killing of K1-encapsulated Escherichia coli. Infect Immun 56:2723–2730
Weiser JN, Gotschlich EC (1991) Outer membrane protein A (OmpA) contributes to serum resistance and pathogenicity of Escherichia coli K-1. Infect Immun 57:2252–2258
Wooster DG, Maruvada R, Blom AM, Prasadarao NV (2006) Logarithmic phase Escherichia coli K1 efficiently avoids serum killing by promoting C4bp-mediated C3b and C4b degradation. Immunol 117:482–493
Acknowledgments
The authors thank Dr. G. Schmidt (Forschungsinstitut Borstel, Germany) for the kind gift of lyophilized samples of E. coli strain no. B 2095 and a sample of K1 A bacteriophage.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution License which permits any use, distribution, and reproduction in any medium, provided the original author(s) and the source are credited.
About this article
Cite this article
Cisowska, A., Bugla-Płoskońska, G. Analysis of the SDS-PAGE patterns of outer membrane proteins from Escherichia coli strains that have lost the ability to form K1 antigen and varied in the susceptibility to normal human serum. Folia Microbiol 59, 37–43 (2014). https://doi.org/10.1007/s12223-013-0262-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12223-013-0262-6