Abstract
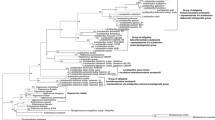
We aimed to clarify if the thermophilic Campylobacter lari organisms including urease-negative (UN) C. lari and urease-positive thermophilic Campylobacter (UPTC) can be differentiated at the species and/or subspecies levels by employing the full-length flaA gene and flaA short variable region (SVR) nucleotide sequence information or not. Thermophilic Campylobacter isolates (n = 45) including UN C. lari (n = 17), UPTC (n = 18), and Campylobacter jejuni (n = 10) were well discriminated at the isolate level by the unweighted pair group method using arithmetic means analysis and neighbor joining procedures constructed based on the full-length flaA gene and flaA SVR nucleotide sequence information. Thus, these procedures may possibly be useful for epidemimological studies for C. lari and C. jejuni.



Similar content being viewed by others
Abbreviations
- MLEE:
-
Multilocus enzyme electrophoresis
- NARTC:
-
Nalidixic acid resistant thermophilic Campylobacter
- PFGE:
-
Pulsed-field gel electrophoresis
- SVR:
-
Short variable region
- UN:
-
Urease-negative (isolates)
- UPTC:
-
Urease-positive thermophilic Campylobacter
References
Benjamin J, Leaper S, Owen RJ, Skirrow MB (1983) Description of Campylobacter laridis, a new species comprising the nalidixic acid resistant thermophilic Campylobacter (NARTC) group. Curr Microbiol 8:231–238
Bolton FJ, Holt AV, Hutchinson DN (1985) Urease-positive thermophilic campylobacters. Lancet I:1217–1218
Debruyne L, On SW, De Brandt E, Vandamme P (2009) Novel Campylobacter lari-like bacteria from humans and molluscs: description of Campylobacter peloridis sp.nov., Campylobacter lari subsp. concheus subsp.nov. and Campylobacter lari subsp. lari subsp.nov. Int J Syst Evol Microbiol 59:1126–1132
Gondo T, Sekizuka T, Manaka N, Murayama O, Millar BC, Moore JE, Matsuda M (2006) Demonstration of the shorter flagellin (flaA) gene of urease-positive thermophilic Campylobacter isolated from the natural environment in Northern Ireland. Folia Microbiol 51:183–190
Lastovica AJ, Skirrow MB (2000) Clinical significance of Campylobacter and related species other than Campylobacter jejuni and C. coli. In: Nachamkin I, Blaser MJ (eds) Campylobacter. ASM Press, Washington, pp 89–120
Lawson AJ, Logan JM, O’Neill GL, Desai M, Stanley J (1999) Large-scale survey of Campylobacter species in human gastroenteritis by PCR and PCR-enzyme-linked immunosorbent assay. J Clin Microbiol 37:3860–3864
Leatherbarrow AJH, Griffiths R, Hart CA, Kemp R, Williams NJ, Diggle PJ, Wright EJ, Sutherst J, Houghton P, French NP (2007) Campylobacter lari: genotype and antibiotic resistance of isolates from cattle, wildlife and water in an area of mixed dairy farmland in the United Kingdom. Environ Microbiol 9:1772–1779
Matsuda M, Moore JE (2004) Urease-positive thermophilic Campylobacter species. Appl Environ Microbiol 70:4415–4418
Matsuda M, Kaneko A, Stanley T, Millar BC, Miyajima M, Murphy PG, Moore JE (2003) Characterization of urease-positive thermophilic Campylobacter subspecies by multilocus enzyme electrophoresis typing. Appl Environ Microbiol 69:3308–3310
Meinersmann RJ, Helsel LO, Fields PI, Hiett KL (1997) Discrimination of Campylobacter jejuni isolates by fla gene sequencing. J Clin Microbiol 35:2810–2814
Meinersmann RJ, Phillips RW, Hiett KL, Fedorka-Cray P (2005) Differentiation of Campylobacter populations as demonstrated by flagellin short variable region sequences. Appl Environ Microbiol 71:6368–6374
Moore JE, Corcoran D, Dooley JSG, Fanning S, Lucey B, Matsuda M, McDowell DA, Megarud F, Millar BC, O’Mahony R, O’Riordan L, O’Rourke M, Rao JR, Rooney PJ, Sails A, Whyte P (2005) Campylobacter Vet Res 36:351–382
Nachamkin I, Bohachick K, Patton CM (1993) Flagellin gene typing of Campylobacter jejuni by restriction fragment length polymorphism analysis. J Clin Microbiol 31:1531–1536
Nuijten PJM, van Asten FJAM, Gasstra W, van der Zeijst BAM (1990) Structural and functional analysis of two Campylobacter jejuni flagellin genes. J Biol Chem 265:17798–17804
Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic tree. Mol Biol Evol 4:406–425
Sekizuka T, Gondo T, Murayama O, Moore JE, Millar BC, Matsuda M (2002) flaA-like sequences containing internal termination codons (TAG) in urease-positive thermophilic Campylobacter isolated in Japan. Lett Appl Microbiol 35:185–189
Sekizuka T, Gondo T, Murayama O, Kato Y, Moore JE, Millar BC, Matsuda M (2004a) Molecular cloning, nucleotide sequencing and characterization of the flagellin gene from isolates of urease-positive thermophilic Campylobacter. Res Microbiol 155:185–191
Sekizuka T, Seki K, Hayakawa T, Moore JE, Murayama O, Matsuda M (2004b) Phenotypic characterisation of flagellin and flagella of urease-positive thermophilic campylobacters. Br J Biomed Sci 61:186–189
Sekizuka T, Yokoi T, Murayama O, Millar BC, Moore JE, Matsuda M (2005) A newly constructed primer pair for the PCR amplification, cloning and sequencing of the flagellin (flaA) gene from isolates of urease-negaive Campylobacter lari. Antonie Leeuwenhoek 88:113–120
Sekizuka T, Murayama O, Moore JE, Millar BC, Matsuda M (2007) Flagellin gene structure of flaA and flaB and adjacent gene loci in urease-positive thermophilic Campylobacter (UPTC). J Basic Microbiol 47:63–73
Thompson JD, Higgins DG, Gibson TJ (1994) CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucl Acids Res 22:4672–4680
Author information
Authors and Affiliations
Corresponding author
Additional information
J. Hirayama and A. Tazumi contributed equally to this manuscript and hence should be considered joint first authors.
Rights and permissions
About this article
Cite this article
Hirayama, J., Tazumi, A., Nakanishi, S. et al. Reliability of nucleotide sequence information of full-length flagellin A gene (flaA) and flaA short variable region (SVR) for molecular discrimination of Campylobacter lari organisms. Folia Microbiol 56, 103–109 (2011). https://doi.org/10.1007/s12223-011-0026-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12223-011-0026-0