Abstract
Fungal infections constitute a serious clinical problem in the group of patients receiving total parenteral nutrition. The majority of species isolated from infections of the total parenteral nutrition patients belong to Candida genus. The most important factors of Candida spp. virulence are the phenomenon of “phenotypic switching,” adhesins, dimorphism of fungal cells and the secretion of hydrolytic enzymes such as proteinases and lipases, including aspartyl proteinases. We determined the proteolytic activity of yeast-like fungal strains cultured from the clinical materials of patients receiving total parenteral nutrition and detected genes encoding aspartyl proteinases in predominant species Candida glabrata—YPS2, YPS4, and YPS6, and Candida albicans—SAP1–3, SAP4, SAP5, and SAP6. C. albicans released proteinases on the various activity levels. All C. glabrata strains obtained from the clinical materials of examined and control groups exhibited secretion of the proteinases. All 13 isolates of C. albicans possessed genes SAP1–3. Gene SAP4 was detected in genome of 11 C. albicans strains, SAP5 in 6, and SAP6 in 11. Twenty-six among 31 of C. glabrata isolates contained YPS2 gene, 21 the YPS4 gene, and 28 the YPS6 gene. We observed that clinical isolates of C. albicans and C. glabrata differed in SAPs and YPSs gene profiles, respectively, and displayed differentiated proteolytic activity. We suppose that different sets of aspartyl proteinases genes as well as various proteinase-activity levels would have the influence on strains virulence.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Fungal infections constitute a serious clinical problem in group of patients after surgical procedures, receiving total parenteral nutrition (TPN). The majority of species isolated from infections of the TPN patients belong to Candida genus. Among the yeast-like fungal strains, Candida albicans is one of the microorganisms that cause infections the most frequently. The other species—Candida parapsilosis, Candida glabrata, and Candida tropicalis—are isolated from clinical materials less commonly (Slodkowski et al. 2004). The most important factors of Candida spp. virulence are the phenomenon of “phenotypic switching”—phenotypic changing, adhesins, dimorphism of fungal cells, and the secretion of hydrolytic enzymes, such as proteinases and lipases, including aspartyl proteinases (Saps). Saps had been recognized as the factors of virulence since the time they were discovered (Tavanti et al. 2004). Some species that are not involved in Sap proteinases secretion may produce enzymes of aspartyl proteinases activity, closely related to Saccharomyces cerevisiae yapsins. All these enzymes, named Yps proteinases, play an important role in C. glabrata virulence (Kaur et al. 2007; Krysan et al. 2005). Some species, including C. albicans, being dimorphic, can grow as yeast cells or in the form of pseudohyphae or hyphae (Wang 2009). This ability is connected with the expression of genes encoding proteinases. It is worth to notice that proteinases are responsible for infection development.
The aim of our study was to determine the proteolytic activity of yeast-like fungal strains cultured from the clinical materials of patients receiving total parenteral nutrition and the detection of genes encoding aspartyl proteinases in predominant species, isolated from examined and control groups. We have also attempt to divide all isolates into more and/or less virulent. The virulence genes for analysis were selected according to their role and importance in development of infections.
Material and methods
The clinical materials consisted of yeast-like 55 fungal strains isolated from 37 patients (hospitalized in Department of Nutrition and Surgery in W. Orlowski Hospital in Warsaw) with TPN from the various clinical materials isolated (Table 1; examined group). In the control group, 35 isolates cultured from the clinical materials of 30 patients without TPN support were analyzed (Table 2).
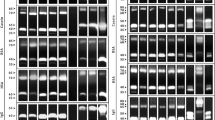
All clinical materials were cultured on Sabouraud agar plates with chloramphenicol and gentamicin (bioMérieux). After a 1- to 2-day incubation at 30°C, all isolates were identified using the ID 32 C test (bioMérieux). The proteolytic activity was determined on modified Staib’s agar (in percent): agar, 2%; glucose, 2%; KH2PO4, 0.1%; and MgSO4, 0.05%, with addition of 0.4 g bovine serum albumin (Biomed) to each 200 mL of the broth (Kurnatowska et al. 1999). For the analysis, the suspensions of 0.5 McFarland Candida cells from 2-day-old Sabouraud agar cultures were prepared. The 10-μL drops of the suspensions were put on the agar surfaces. All plates were incubated up to 7 days at 37°C. The proteolytic zone was observed and evaluated as follows: (++++)—proteolysis after 24 h, (+++)—after 48 h, (++)—after 72 h, (+)—clouding around the colonies, or no reaction (−) after 7 days as described previously by Nowakiewicz and Ziolkowska (2007).
The isolation of genomic DNA was done according to the EURx protocol enclosed to the yeast DNA isolation kit. Three-day-old yeast cultures drown in 1.5 mL Sabouraud liquid broth without any antibiotics addition were used. After removing the broth, the enzymic lysis of the cells was done in buffer containing proteinase K and ribonuclease. All obtained lysates were applied to the binding spin columns and washed twice with buffer to remove lipids and proteins. The elution of DNA bound to the membrane was done with Tris buffer (pH 8.0).
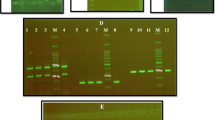
Genotypic analysis was based on the detection of chosen genes encoding fungal proteinases using standard PCR reaction. The analysis was performed for the genes considered to be most important in the development of virulence. The following genes were amplified from the genomic DNA of most isolates: C. glabrata—YPS2, YPS4, and YPS6, and C. albicans—SAP1–3, SAP4, SAP5, and SAP6. The reaction was conducted in DNA Engine thermocycler (MJ Research) under optimized conditions for SAPs genes: 95°C for 5 min, 30 cycles of 95°C for 30 s, 49°C for 45 s, 72°C for 1 min, and 72°C for 5 min; for YPS genes: 95°C for 5 min, 30 cycles 95°C for 1 min, 54°C for 45 s, 72°C for 1 min, and 72°C for −5 min. The method of the amplification generally followed protocols of Kaur et al. (2007) and Kalkanci et al. (2005). Table 3 shows the primer sequences and product lengths for analyzed genes. Reaction products were separated by electrophoresis on 1.5% agarose gel and visualized with ethidium bromide under UV light.
Results
The secretion of proteinases in 19 strains isolated of study group (C. albicans—12, C. parapsilosis—2, C. tropicalis—2, Candida krusei—2, and Candida lusitaniae—1) was observed. In control group, 22 isolates of C. albicans released proteinases on the various activity levels. All C. glabrata strains of examined and control groups secreted proteinases (Table 4).
All 13 isolates of C. albicans in the study group possessed genes SAP1-3. Gene SAP4 was detected in genome of 11 C. albicans strains, SAP5 in 5, and SAP6 in 11 strains. Simultaneous presence of all analyzed genes was detected in case of five C. albicans isolates. Two isolates revealed simultaneous deficiency of SAP5 and SAP6 genes, and two other simultaneously lack the SAP4 and SAP5 genes. In case of four isolates, the absence of SAP5 gene was observed with simultaneous presence of other analyzed genes (see Table 4).
Twenty-six among 31 of C. glabrata isolates in study group possessed YPS2 gene, 21 the YPS4 gene, and 28 the YPS6 gene. The presence of all analyzed genes was demonstrated in case of 21 C. glabrata strains. Three isolates of C. glabrata revealed the lack of YPS2, YPS4, and YPS6 genes, two strains exhibited simultaneous deletion of YPS2 and YPS4. The lack of YPS4 gene with simultaneous presence of other genes was observed in case of five C. glabrata isolates (see Table 4). In control group, all studied SAP genes were detected in genome of 12 strains. Two of them demonstrated lack of the SAP6 gene with simultaneous presence of SAP1–3, SAP4, and SAP5. In case of five isolates, SAP5 gene was absent. Two isolates revealed the deletion of SAP5 and SAP6 genes, one isolate SAP4 and SAP5. The deficiency of SAP1–3, SAP4, and SAP5 genes was observed in case of one strain. Three isolates out of five strains of C. glabrata demonstrated simultaneous lack of YPS2, YPS4, and YPS6 genes. In one isolate, single deletion of YPS6 gene and, in another one, the double deletion of YPS4 and YPS6 were detected.
Discussion
Aspartyl proteinases of Candida albicans are encoded by the multigene family of at least ten different highly regulated genes (SAP1–10) (Tavanti et al. 2004). Those genes were classified into three separated subgroups on base of their amino acid sequences. Enzymes Sap1–3 formed the group with a 75% similarity in sequence whereas Sap4–6 with 90%. Proteinases Sap1–3 play an important role in mucosal infections while Sap4–6 in systemic infections (Felk et al. 2002). Kalkanci et al. (2005) proved that the deletion of SAP4 to SAP6 genes can lead to a virulence reduction and, in consequence, to attenuated development of disseminated fungal infections. C. albicans strains causing mucosal infection are less virulent than the strains that are the etiological factors of candidemia. C. albicans isolates analyzed in our study displayed differences in the gene set of encoded aspartyl proteinases. The detected deletions concerned genes SAP4–6, both in our examined and control groups. The analysis did not reveal mutants in SAP1–3 genes. Considering the literature data, we suggest that strains possessing all analyzed genes (SAP1–3, SAP4, SAP5, SAP6) are able to develop systemic infections, whereas mutants in SAP4–6 genes (as less virulent) would cause infections with milder course.
Protein products of the SAP1–3 genes are secreted by yeast cells and by pseudohyphal form of C. albicans. Sap4–6 proteinases are mostly secreted by pseudohyphae. According to Felk et al. (2002), C. albicans strains that formed pseudohyphal forms but were deprived of accompanied proteinases encoded by SAP6, in particular, demonstrated reduced virulence. The double mutants SAP4–6, SAP5–6, or SAP4–5 were characterized by very similar feature. Our results demonstrated that the majority strains in both examined and control groups possessed deletions of genes from the SAP4–6 subfamily (single or double).
C. albicans is not a unique species of Candida genus secreting proteinases. Other pathogenic Candida, including Candida dubliniensis, C. tropicalis, and C. parapsilosis, possessed also the SAP genes. C. tropicalis has four SAPs, while C. parapsilosis has at least two (Naglik et al. 2003). The existence of cluster of 11 genes encoding aspartyl proteinases (YPS) was showed in the genome of C. glabrata. They play an important role in the maintenance of cell wall integrity, adherence to host cells, and survival of fungal pathogens in macrophages and in virulence. During contact of C. glabrata cells with host macrophages, the transcription of YPS2 gene and other YPS cluster genes, except YPS3 and YPS6, increased. Genes YPS3 and YPS6 were expressed during incubation of yeast cells with macrophages, as well as during Candida growth on agar medium (Kaur et al. 2007). Similarly, we suggest that the strains with deletion of YPS genes could be characterized by modulated virulence level. In our study group, 5 strains of C. glabrata with deletion of YPS2 gene, 10 with deletion of YPS4, and 3 lacking YPS6 gene were detected. However, unambiguous correlation of gene deletions and strain proteolytic activities were not observed. C. albicans species displayed different proteolytic activity, and the majority of isolates secreted high levels of enzymes (Kurnatowska et al. 1999). On the other hand, in case of C. glabrata isolates, the activity was low or even very low, which agrees with the findings of Li et al. (2007) who suggested that C. glabrata exhibited extracellular proteinase activity but not on significant levels. The differences can be explained, e.g., by distinctive functions of corresponding enzyme in both species.
In conclusion, we confirmed the heterogeneity of gene encoding proteinases in C. albicans and C. glabrata clinical isolates and different SAPs and YPSs gene profiles, which can affect the strain virulence.
Abbreviations
- PCR:
-
Polymerase chain reaction
- SAP:
-
Secreted aspartyl proteinase gene
- Saps:
-
Secreted aspartyl proteinases
- TPN:
-
Total parenteral nutrition
- YPS:
-
Yapsin-related aspartyl proteinase gene
References
Felk A, Kretschmar M, Albrecht A, Schaller M, Beinhauer S, Nichterlein T, Sanglard D, Korting HC, Schäfer W, Hube B (2002) Candida albicans hyphal formation and the expression of the EFG-1-regulated proteinases SAP4 to SAP6 are required for the invasion of parenchymal organs. Infect Immun 70:3689–3700
Kalkanci A, Bozdayi G, Biri A, Kustimur S (2005) Distribution of secreted aspartyl proteinases using a polymerase chain reaction assay with SAP specific primers in Candida albicans isolates. Folia Microbiol 50:409–413
Kaur R, Ma B, Cormack BP (2007) A family of glycosylphosphatidylinositol-linked aspartyl proteases is required for virulence of Candida glabrata. Proc Natl Acad Sci USA 104:7628–7633
Krysan DJ, Ting EL, Abeijon C, Kroos L, Fuller RS (2005) Yapsins are a family of aspartyl proteases required for cell wall integrity in Saccharomyces cerevisiae. Eukaryot Cell 4:1364–1374
Kurnatowska AJ, Rozga A, Kurnatowski P (1999) Activity of aspartic proteinase of fungal oral isolates. (In Polish) Mikol Lek 6:21–25
Li L, Redding S, Dongari-Bagtzohlou A (2007) Candida glabrata, an emerging oral opportunistic pathogen. J Dent Res 86:204–215
Naglik RJ, Challacombe SJ, Hube B (2003) Candida albicans secreted aspartyl proteinases in virulence and pathogenesis. Microbiol Mol Biol Rev 3:400–428
Nowakiewicz A, Ziolkowska G (2007) Phenotypic characteristics of Malassezia pachydermatis strains. (In Polish) Medycyna Wet 63:448–454
Slodkowski M, Cebulski W, Deptala A, Krasnodebski IW (2004) Fungal infections in patients receiving parenteral and enteral nutrition. (In Polish) Zakazenia 4:45–49
Tavanti A, Pardini G, Campa D, Davini P, Lupetti A, Senesi S (2004) Differential expression of secretory aspartyl proteinase genes (SAP-10) in oral Candida albicans isolates with distinct karyotypes. J Clin Microbiol 10:4726–4734
Wang Y (2009) CDKs and the yeast–hyphal decision. Curr Opin Microbiol 12:644–649
Acknowledgments
This work was supported by the Polish State Committee for Scientific Research (grant no. NN404 093735).
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
Open Access This is an open access article distributed under the terms of the Creative Commons Attribution Noncommercial License (https://creativecommons.org/licenses/by-nc/2.0), which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
About this article
Cite this article
Sikora, M., Dabkowska, M., Swoboda-Kopec, E. et al. Differences in proteolytic activity and gene profiles of fungal strains isolated from the total parenteral nutrition patients. Folia Microbiol 56, 143–148 (2011). https://doi.org/10.1007/s12223-011-0023-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12223-011-0023-3