Abstract
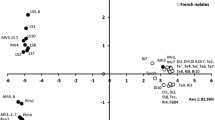
Genetic variability of Phoma sorghina, a ubiquitous facultative phytopathogen, was investigated on 41 isolates cultivated from surface-sterilized sorghum grains originating from South Africa and Texas; pearl millet isolates from Namibia were also included. Most of the isolates from Texas produced intense red pigments, especially on Czapek-Dox agar plates. Many African isolates formed conspicuous dark radial substrate hyphae with intercalated chlamydospores on oatmeal plates. Conidial dimensions and shape were very variable (mean lengths 4.5–5.7 μm). Haplotypes were defined based on 53 markers from banding patterns obtained with rep-PCR (primers: M13core, ERIC IR). The shared geographic origin was partially reflected in the clades of the haplotype phylogram. The values of G ST were intermediate; 16–37 % of the variation was found between the populations. Nm values of gene flow were 0.84–1.15. Average gene diversity H E was moderate (0.256). Sequences of ITS-rDNA were obtained from 21 isolates. Allele 1 was found in 9 isolates scattered throughout the clades, allele 2 occurred in 6 isolates (5 of them from the same clade), alleles 3 and 4 were shared by two isolates each and two isolates were unique. Alleles 1 and 2 were also found among highly related sequences from GenBank. All shared an 8-bp deletion near the 5′ end of ITS2 that was not found in any other Phoma/Didymella species and which may be a typical marker for P. sorghina. Among related species, members of legume-associated Ascochyta/Didymella complex, Epicoccum spp., D. applanata and P. glomerata were found.
Similar content being viewed by others
Abbreviations
- AFLP:
-
amplified fragment length polymorphism
- NNI:
-
nearest neighbor interchange (method)
- ERIC:
-
repetitive intergenic consensus (sequence)
- OA:
-
oatmeal agar
- GTR:
-
general time reversible (substitution model)
- PCR:
-
polymerase chain reaction
- ITS:
-
internal transcribed spacer
- RAPD:
-
random amplification of polymorphic DNA
- MEA:
-
2 % malt extract, 2 % agar (medium)
- UPGMA:
-
unweighted pair group cluster method with arithmetic averages
References
Abeln E.C.A., Stax A.M., DE Gruyter J., VAN DER Aa H.A.: Genetic differentiation of Phoma exigua varieties by means of AFLP fingerprints. Mycol.Res. 106, 419–427 (2002).
Abler S.W.: Ecology and Taxonomy of Leptosphaerulina spp. Associated with Turfgrasses in the United States. Virginia Polytechnic Institute and State University, Blacksburg (USA) 2003.
Amaral A.L.D., Carli M.L.D., Neto J.F.B., Soglio F.K.D.: Phoma sorghina, a new pathogen associated with phaeosphaeria leaf spot on maize in Brazil. Plant Pathol. 53, 259 (2004).
Anahosur K.H., Sivanesan A.: Mycosphaerella holci. CMI Descriptions of Fungi and Bacteria (No. 59). Sheet 584 (1978).
Arenal F., Platas G., Monte E., Pelaez F.: ITS sequencing support for Epicoccum nigrum and Phoma epicoccina being the same biological species. Mycol.Res. 104, 301–303 (2000).
Arora D.K., Hirsch P.R., Kerry B.R.: PCR-based molecular discrimination of Verticillium chlamydosporium isolates. Mycol.Res. 100, 801–809 (1996).
von Arx J.A.: Plant pathogenic fungi. Nova Hedwigia Beihefte 87, 1–288 (1987).
Aveskamp M.M., De Gruyter J., Crous P.W.: Biology and recent developments in the systematics of Phoma, a complex genus of major quarantine significance. Fungal Diversity 31, 1–18 (2008).
Boerema G.H., DE Gruyter J., Noordeloos M.E., Hamers M.E.C.: Phoma Identification Manual: Differentiation of Specific and Infra-Specific Taxa in Culture. CABI Publishing, Wallingford (UK) 2004.
Castell-Miller C.V., Szabo L.J., Gale L.R., O’Neill N.R., Samac D.A.: Molecular variability of a Minnesota population of Phoma medicaginis var. medicaginis, the causal agent of spring black stem and leaf spot of alfalfa. Can.J.Plant Pathol. 30, 85–96 (2008).
Excoffier L., Laval G., Schneider S.: Arlequin ver. 3.0: an integrated software package for population genetics data analysis. Evol. Bioinf.Online 1, 47–50 (2005).
Faris Mokaiesh S., Boccara M., Denis J.B., Derrien A., Spire D.: Differentiation of the “Ascochyta complex” fungi of pea by biochemical and molecular markers. Curr.Genet. 29, 182–190 (1996).
Feldman T.S., O’Brien H.E., Arnold A.E.: Moths that vector a plant pathogen also transport endophytic fungi and mycoparasitic antagonists. Microb.Ecol. 56, 742–750 (2008).
Felsenstein J.: Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39, 783–791 (1985).
Forbes G.A., Bandyopadhyay R., Garcia G.: A review of sorghum grain mold, in W.A.J. DE Milliano, R.A. Frederiksen, G.D. Bengston, Eds): Sorghum and Millets Diseases: a Second World Review. ICRISAT, Pantacheru (Andhra Pradesh, India) 1992.
Goodwin S.B., Dunkle L.D., Zismann V.L.: Phylogenetic analysis of Cercospora and Mycosphaerella based on the internal transcribed spacer region of ribosomal DNA. Phytopathology 91, 648–658 (2001).
de Gruyter J., Aveskamp M.M., Woudenberg J.H.C., Verkley G.J.M., Groenewald J.Z., Crous P.W.: Molecular phylogeny of Phoma and allied anamorph genera: towards a reclassification of the Phoma complex. Mycol.Res. 113, 508–519 (2009).
Guindon S., Gascuel O.: A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst.Biol. 52, 696–704 (2003).
Guindon S., Lethiec F., Duroux P., Gascuel O.: PHYML Online — a web server for fast maximum likelihood-based phylogenetic inference. Nucl.Acids Res. 33, W557–W559 (2005).
Gure A.: Seed-Borne Fungi of the Afromontane Tree Species Podocarpus falcatus and Prunus africana in Ethiopia. PhD Thesis. Swedish University of Agricultural Sciences, Uppsala 2004.
Hall T.A.: BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucl.Acids Symp.Ser. 41, 95–98 (1999).
Hampl V., Pavlíček A., Flégr J.: Construction and bootstrap analysis of DNA fingerprinting-based phylogenetic trees with a freeware program FreeTree: application to trichomonad parasites. Internat.J.Syst.Evol.Microbiol. 51, 731–735 (2001).
Healy A., Reece K., Walton D., Huong J., Shah K., Kontoyiannis D.P.: Identification to the species level and differentiation between strains of Aspergillus clinical isolates by automated repetitive-sequence-based PCR. J.Clin.Microbiol. 42, 4016–4024 (2004).
Healy M., Reece K., Walton D., Huong J., Frye S., Raad I.I., Kontoyiannis D.P.: Use of the DiversiLab System for species and strain differentiation of Fusarium species isolates. J.Clin.Microbiol. 43, 5278–5280 (2005).
Kretzer A., Li Y., Szaro T.M., Bruns T.D.: Internal transcribed spacer sequences from 38 recognized species of Suillus sensu lato: phylogenetic and taxonomic implications. Mycologia 88, 776–785 (1996).
Lewontin R.C.: The apportionment of human diversity. Evolutionary Biol. 6, 381–398 (1972).
Lieckfeldt E., Meyer W., Börner T.: Rapid identification and differentiation of yeasts by DNA and PCR fingerprinting. J.Basic Microbiol. 33, 413–426 (1993).
Lindqvist-Kreuze H., Hellqvist S., Koponen H., Valkonen J.P.T.: Phoma-Didymella complex on hybrid arctic bramble with wilting symptoms. Plant Pathol. 52, 567–578 (2003).
Mahdi L.: A Survey of Hawaiian Marine Fungi and Yeast. MSc Thesis. University of Hawai’i, Manoa 2006.
Maynard Smith J., Feil E.J., Smith N.H.: Population structure and evolutionary dynamics of pathogenic bacteria. Bioessays 22, 1115–1122 (2000).
Menkis A., Vasiliauskas R., Taylor A.F.S., Stenstrom E., Stenlid J., Finlay R.: Fungi in decayed roots of conifer seedlings in forest nurseries, afforested clear-cuts and abandoned farmland. Plant Pathol. 55, 117–129 (2006).
Nei M.: Analysis of gene diversity in subdivided populations. Proc.Nat.Acad.Sci.USA 70, 3321–3323 (1973).
Nei M.: Molecular Evolutionary Genetics. Columbia University Press, New York 1987.
Pažoutová S., Kolínská R.: Cerebella relationship to Epicoccum and their closest relatives among Dothideales. Czech Mycol. 54, 155–160 (2003).
Peever T.L., Barve M.P., Stone L.J., Kaiser W.J.: Evolutionary relationships among Ascochyta species infecting wild and cultivated hosts in the legume tribes Cicereae and Vicieae. Mycologia 99, 59–77 (2007).
Pethybridge S.J., Scott J.B., Hay F.S.: Genetic relationships among isolates of Phoma ligulicola from pyrethrum and chrysanthemum based on ITS sequences and its detection by PCR. Australas.Plant Pathol. 33, 173–181 (2004).
Rabie C.J., van Rensburg S.J., van der Watt J.J., Lubben A.: Onyalai — the possible involvement of a mycotoxin produced by Phoma sorghina in the aetiology. South Afr.Med.J. 57, 1647–1650 (1975).
Reddy P.V., Patel R., White J.F. Jr.: Phylogenetic and developmental evidence supporting reclassification of cruciferous pathogens Phoma lingam and Phoma wasabiae in Plenodomus. Can.J.Bot. 76, 1916–1922 (1998).
Simon U.K., Weiss M.: Intragenomic variation of fungal ribosomal genes is higher than previously thought. Mol.Biol.Evol. 25, 2251–2254 (2008).
Somai B.M., Dean R.A., Farnham M.W., Zitter T.A., Keinath A.P.: Internal transcribed spacer regions 1 and 2 and random amplified polymorphic DNA analysis of Didymella bryoniae and related Phoma species isolated from cucurbits. Phytopathology 92, 997–1004 (2002).
De Souza N., Zambolim L., Thièbaut J.T.L.: Variabilidade de isolados de Phoma sorghina em arroz. Pesq.Agropec.Bras. 23, 1139–1141 (1988).
de Souza Borges W., Tallarico Pupo M.: Novel anthraquinone derivatives produced by Phoma sorghina, an endophyte found in association with the medicinal plant Tithonia diversifolia (Asteraceae). J.Braz.Chem.Soc. 17, 929–934 (2006).
Sullivan R.F., White J.F.: Phoma glomerata as a mycoparasite of powdery mildew. Appl.Environ.Microbiol. 66, 425–427 (2000).
Tymon A.M., Pell J.K.: ISSR, ERIC and RAPD techniques to detect genetic diversity in the aphid pathogen Pandora neoaphidis. Mycol.Res. 109, 285–293 (2005).
Venkatasubbaiah P., Van Dyke C., Chilton W.: Phytotoxic metabolites of Phoma sorghina, a new foliar pathogen of pokeweed. Mycologia 84, 715–723 (1992).
Verkley G.J.M., Starink-Willemse M., van Iperen A., Abeln E.C.A.: Phylogenetic analyses of Septoria species based on the ITS and LSU-D2 regions of nuclear ribosomal DNA. Mycologia 96, 558–571 (2004).
Versalovic J., Koeuth T., Lupski R.: Distribution of repetitive DNA sequences in eubacteria and application to fingerpriting of bacterial genomes. Nucl.Acids Res. 19, 6823–6831 (1991).
Waldrop M.P., Zak D.R., Blackwood C., Curtis C.D., Tilman D.: Resource availability controls fungal diversity across a plant diversity gradient. Ecol.Lett. 9, 1127–1135 (2006).
Wang G., Li Q., Zhu P.: Phylogenetic diversity of culturable fungi associated with the Hawaiian sponges Suberites zeteki and Gelliodes fibrosa. Antonie van Leeuwenhoek 93, 163–174 (2008).
White J.F. Jr., Morgan-Jones G.: Studies in the genus Phoma. II. Concerning Phoma sorghina. Mycotaxon 18, 5–13 (1983).
White T.J., Bruns T., Lee S., Taylor J.: Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics, pp. 315–322 in M.A. Innis, D.H. Gelfand, J.J. Sninsky, T.J. White (Eds): PCR Protocols: a Guide to Methods and Applications. Academic Press, San Diego (USA) 1990.
Wright S.: Evolution in Mendelian populations. Genet.Mol.Biol. 16, 97–159 (1931).
Yeh F.C., Yang R., Boyle T.J., Ye Z., Xiyan J.M.: PopGene32, Microsoft Windows-Based Freeware for Population Genetic Analysis. 1.32. Molecular Biology and Biotechnology Centre, University of Alberta, Edmonton (Alberta, Canada) 2000.
Yoshioka K.: KyPlot — a user-oriented tool for statistical data analysis and visualization. Comp.Stat. 17, 425–437 (2002).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Pažoutová, S. Genetic variation of Phoma sorghina isolates from Southern Africa and Texas. Folia Microbiol 54, 217–229 (2009). https://doi.org/10.1007/s12223-009-0035-4
Received:
Revised:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12223-009-0035-4