Abstract
This paper presents the results of genetic variability analyses using genealogical data on the main genetic bloodline of the Australian Cattle Dog in Italy, a line that has had a significant impact on the development of the breed. All the genealogical data on the progeny and ancestors of one of the first stallions introduced in Italy were considered, i.e. Cattlefarm's Comeback Jack born on 1/2/1997 in Finland. Animals from the bloodline born between 1962 and 2019 were considered. A total number of 1722 animals were found to be from the line which represents the entire population (WP), including the basic population (BP) and the reference population (RP) defined as the animals currently living. A total of 982 animals were in the RP, with the oldest living dogs born in 2004. A total of 854 dogs were inbred. The average inbreeding coefficient (F) in the RP was 5.1%, while the average inbreeding of the inbred animals was 5.8%. The F was < 0.10 in 711 dogs (77.3% of inbred), and > 0.20 in only 36 dogs (3.91% of inbred). Fifteen traced generations were highlighted. A maximum average inbreeding value (6.45%) was observed in the dogs with 11 traced generations. This research highlighted the good genetic variability of this Australian Cattle Dog bloodline thanks to the efficient management of the breeders who in the past introduced some stallions from abroad. Currently, the lines in Italy are not sufficiently high to prevent inbreeding in the new matings, which is becoming frequent. It is, therefore, important to continue to import new stallions for reproduction to expand the genetic variability. However, at the same time, the old lines need to be preserved genetically, aptitudinally and morphologically, as they are an important heritage of the breed in Italy.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
The Australian Cattle Dog originated in Australia, descended from Halls Heelers, and is a dog produced from crossing the Northumberland Blue Merle Drovers dogs with dingos (Clark 2003a). It is possible that this dog may carry some dingo alleles and falls closer to the dingo reference group than other dog breeds, such as Labrador Retrievers or Doberman Pinschers (Elledge et al. 2006). Some dog breeders also believe that Dalmatian and Australian Kelpies were included in the breeding of the Australian Cattle Dog (Clark 2003b). Bull Terrier infusions may also have occurred (Sanderson 1981). The Australian Cattle Dog breed belongs to Group 1 (sheepdogs and cattle dogs—except Swiss Cattle Dogs), Sect. 2 (cattle dogs—except Swiss cattle dogs) of the Fédération Cynologique Internationale (FCI) and of the Ente Nazionale Cinofilia Italiana (ENCI). In Italy, the official breed Club (the Australian Cattle Dog Club Italia 2021) was founded and recognized by the ENCI in 2015. The Australian Cattle Dog was bred to help spread cattle farming in old Australia. It is a strong, compact, harmoniously built working dog that has the ability and desire to fulfill the task it has been given. Its substance, power, balance of its structure and the quality of its muscles contribute to the impression of great agility, strength and endurance (Libro Genealogico ENCI 2021). The Australian Cattle Dog is born to lead cattle; however, in recent years, it has participated in a range of activities beyond herding, including competing in sporting events and working as assistance dogs. It has been used in utility and defense tests with excellent results. Its temper, agility and "easy” bite made it an excellent dog in IPO tests (formerly known as Schutzhund; IPO stands for Internationale Prüfungs-Ordnung). IPO is a part sport which includes tracking, obedience and protection, and is common in northern Europe and Australia. Dogs of any breed, even mixes, compete in IPOs, however the most common breeds are German Shepherds, Belgian Malinois, Boxers, Rottweilers, Dobermans, Cane Corso, Giant Schnauzers, Bouvier des Flandres, Dutch Shepherd Dogs, Beaucerons, American Bulldogs, Black Russian Terriers, and Airedale Terriers (DVG 2021).
The Standard (FCI Standard No. 287/05.09.2013) reports a height at the withers of about 46–51 cm for males, and 43–48 cm for females. The coat colors permitted include two typical colors, red or black and tan (‘blue’) with varying degrees of mottling and/or speckling (Fig. 1). Blue dogs can be blue, blue mottled, or blue speckled always with tan and with or without black markings. Red dogs are evenly speckled with solid red markings. Both red dogs and blue dogs are born white (except for any solid-colored body or face markings) and the red or black hairs grow as they mature. The distinctive adult coloration is the result of black or red hairs closely interspersed through a predominantly white coat. This is not merle coloration (a speckled effect that has associated health issues), but rather the result of the ticking gene (Kennel Working Mates 2021).
The variation in basecoat color is most likely controlled by the dominant ay and recessive at alleles at the agouti (A) locus or ASIP gene (Berryere et al. 2005). Occasionally cream-colored dogs are born from red or blue parents. The cream color does not correspond to the breed standard, and cream-colored dogs cannot be registered (Dürig et al. 2018).
Dogs that are the result of dingos mating with dingo-like domestic dogs, such as the Australian Cattle Dog and Australian kelpie, may display dingo-like coloration and body form (Corbett 1995, 2001; Fleming et al. 2001). Corbett (1995) reported that ticking was often observed in F1 hybrids of dingoes and Australian Cattle Dogs (Elledge et al. 2006).
The most common health problems, all controlled and tested by ENCI for registration of the dogs in the Selected Reproduction, are congenital hereditary sensorineural deafness and progressive retinal atrophy, primary lens luxation, hip dysplasia and elbow dysplasia; otherwise, it is a robust breed with a lifespan of 12–14 years. Lee (2011) reported 13.41 ± 2.36 years as the average value with an extreme longevity of up to 30 years (Mazzatenta et al. 2017).
In Italy, where our research was carried out, approximately 290 puppies are registered every year, and there are only 20 official breeders distributed in eight regions in the center-north (average value 2.5 ± 1.31 breeders per region) and particularly in Lazio and Piemonte (four breeders per region) (ENCI 2021).
Line-breeding and selection in dogs have successfully led to desirable phenotypic traits. However, they have also had the unintended consequence of increasing inbreeding, which in turn can increase genetic monofactorial and multifactorial diseases (Cecchi et al. 2020) and lead to inbreeding depression (Ólafsdóttir and Kristjánsson 2006) on fitness (Brzeski et al. 2014; Cecchi et al. 2016a), production (Leroy 2014) and morphological traits (Gandini et al. 1992; Gómez et al. 2009; Wolc and Baliñska 2010; Cecchi et al. 2018).
Given the limited number of animals born in Italy and the subsequent risk of increased inbreeding, the aim of the present work was to verify the management of matings by breeders. For the first time, we evaluated the genetic variability of the oldest Australian Cattle Dog bloodline in Italy, a line that has had a significant impact on the development of the breed in Italy (the first stallion was Cattlefarm's Comeback Jack born on 1/2/1997 in Finland, blue).
2 Materials and methods
2.1 Sample collection
The pedigree of the dogs born between 1962 and 2019 was considered. All dogs of the bloodline constitute the whole population (WP) containing all founders, ancestors, and their offspring. The population is divided into the base population (BP) defined as individuals with one or both unknown parents, and the reference population (RP) defined as the population of interest that includes living reproductive animals. Demographic data were collected from the Libro Genealogico ENCI (2021).
2.2 Statistical analysis
We used Pedigree Viewer software (Kinghorn 1994) to verify whether there were any errors left in the data files and also to show the full pedigree structure of the population. The other analyses were carried out with ENDOG software, developed by Gutiérrez and Goyache (2005), and with CFC software (Sargolzaei et al. 2006).
The parameters considered were:
-
Pedigree completeness Pedigree completeness expresses how much generation equivalent information there is on average in any individual’s pedigree. The number of maximum generations known and number of full generations known was also calculated. Pedigree completeness determines how accurately the inbreeding coefficient can be estimated, and therefore has a strong influence on the validity of the estimated values.
-
Popular sires and dams Popular sires were defined as sires with a number of recorded offspring greater than 40, while popular dams were defined as dams with a number of recorded offspring higher than 30 offspring.
-
Generation interval (L). The generation interval is defined as the average age of the parents, when their offspring that were used in breeding were born. This parameter was estimated for the whole pedigree, and also for the reference population separately. In the current study, generation intervals were estimated on two different paths separately: sire–son and dam–daughter. The average generation interval value was computed considering four paths (sire–son, sire-daughter, dam-son and dam–daughter).
-
Inbreeding coefficient (F) The inbreeding coefficient is the probability that any randomly drawn locus of a given individual has two identical by descent alleles (Wright 1922). This parameter was calculated by the tabular method described by Meuwissen and Luo (1992). The number of inbred and the average inbreeding coefficient per sex and in animals divided into those alive and dead were performed using CFC software (Sargolzaei et al. 2006). The distribution of the inbreeding level in the whole population was analyzed, and eight different class levels of inbreeding were considered: 0 < F ≤ 0.05; 0.05 < F ≤ 0.10; 0.10 < F ≤ 0.15; 0.15 < F ≤ 0.20; 0.20 < F ≤ 0.25; 0.25 < F ≤ 0.30; 0.30 < F ≤ 0.35; 0.35 < F ≤ 0.40 (Sargolzaei et al. 2006). The inbreeding coefficients (F), the number of inbred animals and average inbreeding coefficient and the mean AR for each traced generation were also computed (ENDOG software, Gutiérrez and Goyache 2005).
-
Rate of inbreeding (DF) The rate of inbreeding was calculated to follow the change in inbreeding over time and was expressed per unit of time (generations, years, etc.). In the current study, the rate of inbreeding was calculated per generation for the whole population and for the reference population. Individuals that were present in more than one generation were included in all those generations in which they were present.
-
Effective population size (Ne) The effective population size is the number of animals that would lead to the same increase in inbreeding, as observed in the population under study, if they contributed equally to the next generation.
-
Average relatedness (AR) The average relatedness gives the probability that a randomly drawn allele from the population belongs to a given individual, or to a group of individuals. The average relatedness expresses how an individual or a group of individuals represents the population. The average relatedness is an appropriate measure of inbreeding in the case of panmixia. Differences between average inbreeding and average relatedness are therefore informative of the nonrandom mating structure of the population (Goyache et al. 2003). The average relatedness was calculated per generation from the individual average relatedness values. The generations were determined based on the pedigree of the reference population. To prevent bias during the calculation of AR in each generation, individuals that were present in more than one generation were included in all those generations in which they were present.
3 Results and discussion
3.1 Demographic data
From 2005 to 2008, only 167 dogs were recorded (average value 41.7 ± 19.26 per year). Starting from 2009, an increase in enrollments was observed with an average value of 258.6 ± 98.74 dogs per year (2009–2019) with the highest value in 2016 (390 recorded).
The complete database (WP) resulted in 1722 records, with the oldest animals born in 1964 and the youngest born in 2019. Figure 2 shows the pedigree quality in this population of Australian Cattle Dogs. The percentage of completeness was computed as about 81% for the parental generation, from 70.1 to 71.8% for the grandparent generation, and from 63.8 to 65.3% for the third-generation of great-grandparents. The pedigree completeness level was similar both in the dam and sire pathway.
The mean maximum generations and the mean complete generations were 7.61 and 3.12 respectively, while the number of equivalent generations traced (the total of all the generations of ancestors, the ancestors’ proportion known to every generation) was 4.74. The average generation interval for animals of the RP was 3.3 ± 0.40 for stallions and 5.1 ± 0.98 for dams, with an average value of 3.6 ± 0.29. This value is consistent with that reported by Shariflou et al. (2011) in the same breed reared in Australia, but reported for a larger number of animals (74,290 registered dogs). The higher value in females is due to the fact that they are made to reproduce later than males because it is a good practice for breeders to mate females in the second useful heat (Pagnacco 2004), thus extending the generation interval. The male value was lower than that observed for Barbet and Berger des Pyrénées, Epagneul Breton and Montagne des Pyrénées and Beauceron dog breeds (Leroy et al. 2006), for Italian Braque (Cecchi et al. 2013), Braque Saint-Germain (Leroy et al. 2006), and Braque Français type Pyrénées (Cecchi et al. 2016b). On the other hand, the values found for the females are in line with or even higher than those reported in the literature for other breeds. The oldest dog in the RP was born in 2004 (15.8 years old), while the youngest was born in 2019 (7 months old) with an average age of the population of 8.3 ± 2.74.
3.2 Genealogical data and popular sires and dams
A total of 828 males and 894 females were recorded, while 420 sires and 499 dams (Table 1) were recorded, with a dams–sires ratio of 1.19. This value was lower than that reported for the Braque Francais type Pyrenees reared in Italy (1.31; Cecchi et al. 2016b) and in the Italian Braque (1.63; Cecchi et al. 2013), showing a better ratio between sexes according to conservation genetics (Frankham et al. 2002).
Only four popular sires were recorded (0.95% of the sires), and two popular dams (0.40% of the dams). Among the four popular sires, a significant impact on the breed development was exerted by Blackeyed Jack (LO0934058; born on 16/05/2008) with 43 sons. This sire presents a genealogy based on the first Italian stallions and dams such as Cattlefam’s Comeback Jack, Princess 2, Rambo 2, Ol’man Drover’s Boy, and was paired with only two dams, one of which is one of the two popular dams—Aussie Blue Daniels, a female of Australian origin used to introduce new blood in the first lines in Italy. The offspring of these dogs had a large impact on the populations of the Australian Cattle Dog reared in Italy.
A significant impact on the breed development in Italy was also exerted by the stallion Armani Kazari Toyo-Ken (LO08392; born on 29/12/2006), first paired in 2007, with 42 sons resulting from the coupling of eight different dams. Despite being imported from the Czech Republic, and therefore not a direct descendant of Cattlefam's Comeback Jack, this stallion has been frequently used to increase the genetic variability of the breed.
In addition to the number of sires and dams, Table 1 summarizes the other main genealogical parameters computed by ENDOG. A total of 982 animals were in the reference population (477 males and 505 females), 86.96% of which were inbred. The number of animals in the base population (one or both unknown parents: absolute founders = ft) was 306 (17.77%).
A total of 232 ancestors were in the RP, while the effective number of ancestors (fa) was 24. To explain 50% of the genetic variability, a total of 10 ancestors enough were highlighted. These results reveal that only a few males were used in reproduction.
3.3 Inbreeding coefficient
The average inbreeding value (F) and average numerator relationship (AR) in the whole population were 3.23% and 4.76%, respectively. The average F and AR values in the reference population (RP) were 5.08% and 11.83%, respectively. No differences were observed between males and females in the WP and RP.
Many studies, including several from our research group, have reported inbreeding results for Italian or foreign breeds. These cover the entire genealogical database of the breed, such as the study by Cecchi et al. (2013) on the Italian Braque, or by Cecchi et al. (2016a, b) on the Braque Francais type Pyrenees or small populations attributable to individual farms, such as the research carried out on the Pitt Bull (Ciampolini et al. 2013), on the Cocker Spaniel (Cecchi et al, 2020) or a few farms of a certain breed such as the study carried out on the Basset Hound (Cecchi et al. 2018).
Obviously, where there are genealogical data on an entire breed, the results are expected to be lower in terms of inbreeding, whereas with small populations, the coefficients are expected to be higher. In reality, this is not always true, and it all depends on the sensitivity regarding problems attributable to the inbreeding of individual breeders and breed clubs.
The results are, therefore, very variable, for example, in a study on breeds raised in France (Leroy et al. 2006), the average coefficient of inbreeding ranged from 3.3% in Bouledougue Francais to 12.4% in the Barbet. In a genealogical survey of Australian registered dog breeds (Shariflou et al. 2011), the mean inbreeding coefficients ranged from 0% (Central Asian shepherd) to 10.1% (Bichon Frise). Cole et al. (2004) reported that populations of German Shepherd and Labrador Retriever guide dogs exhibited an average inbreeding level of 26.2% and 22% respectively, while in an Italian colony of guide dogs, Cecchi et al. (2009) reported values of 0.45% in German Shepherds, 0.38% in Labradors, and 0.49% for Golden Retrievers. In an Italian population of Pit Bull Terriers, the average inbreeding was low (3.73%; Ciampolini et al. 2013), as was the average inbreeding in the Braque Francais type Pyrenees reared in Italy (4.35%; Cecchi et al. 2016b).
Our value of 5.08% observed in the RP is good given that we analyzed the genetic variability of a single line of the Australian Cattle Dog. In addition, this value is in line with Shariflou et al. (2011) in the same breed but reared in Australia (F = 5.7%).
Considering, therefore, the complete database, 920 dogs were recorded as inbred (53.43%); 711 dogs of which (77.28% of the inbred), showed an inbreeding value lower than 10% which is in line with Shariflou et al. (2011) in the same breed reared in Australia (78% of inbred). Thirty-six dogs (3.91% of the inbred) showed an inbreeding value higher than 20% (Table 2).
The average increase in inbreeding was estimated to be 0.43% per generation for the WP (Ne = 116.30), and 0.86% (Ne = 67.86) for the RP, similarly to the data reported by Shariflou et al. (2011) in the same breed. The inbreeding trend increased with rather low values in comparison to those reported in other breeds with higher numbers in Italy such as the Bolognese (Sabbioni et al. 2008) and Italian Braque (Cecchi et al. 2013). The effective population size, a key parameter that strongly influences the nature and rate of genetic information transmission across generations, population variability and conservation strategies, was more than 50 and thus confirms the good genetic variability of the population (Harmon and Braude 2010).
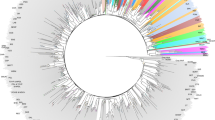
Figure 3 shows the whole genealogy of the population. The population is stratified into 16 traced generations, where the first line represents the base population (generation 0), and the last line reports the animals of the sixteenth generation.
Population statistics on average inbreeding are reported in Table 3. The percentage of inbred increased with the increase in the number of traced generations from 2 to 6. There were few inbred animals (max 36%) but with a very high average inbreeding (up to about 13%). Beginning with the subjects with 12 traced generations, more than 95% of the animals were inbred with an average inbreeding value ranging from 4.2 to 6.2%. With the exception of animals with 11 traced generations, all the average inbreeding coefficients in the other tracked generations were below 6.25%, i.e. the value resulting from the mating of two animals sharing two grandparents (cousin mating). The animals with 15 traced generations were all inbred.
4 Conclusions
The level of genetic variability in our Australian Cattle population was good because only some breeders used closed inbreeding matings, between dogs that basically derived from the first imported dogs to fix the characteristics of the breed. Over the generations matings with the inbreeding method have increased, however, paying close attention to the kinship between the animals. A few years after the arrival of the Australian Cattle Dog in Italy, some breeders decided to introduce animals from abroad into the line, thus limiting inbreeding.
Nevertheless, the lines and the total number of animals bred in Italy are sufficiently high to be able to prevent inbreeding in the latest matings, as highlighted by the fact that all the dogs in the last generation were inbred. It is thus important to import new subjects for reproduction to continue to maintain and/or expand the current genetic variability in breeds. At the same time, it is necessary to pay attention to the matings to genetically, attitudinally and morphologically preserve the old lines, which are an important heritage of the breed in Italy.
References
Australian Cattle Dog Club Italia (2021) Unico Club in Italia per la tutela della razza dal 2015 ufficialmente riconosciuto ENCI. https://www.acdclubitalia.it/ Accessed 28 Jan 2021
Berryere TG, Kerns JA, Barsh GS, Schmutz SM (2005) Association of an Agouti allele with fawn or sable coat color in domestic dogs. Mamm Genome 16:262–272
Brzeski KE, Rabon DRJ, Chamberlain MJ, Waits LP, Taylor SS (2014) Inbreeding and inbreeding depression in endangered red wolves (Canis rufus). Mol Ecol 23:4241–4255
Cecchi F, Bramante A, Mazzanti E, Ciampolini R (2009) A colony of dog guides: analysis of the genetic variability assessed by pedigree data. Ital J Anim Sci 8:48–50
Cecchi F, Paci G, Spaterna A, Ciampolini R (2013) Genetic variability in Bracco Italiano dog breed assessed by pedigree data. Ital J Anim Sci 12:348–352
Cecchi F, Giacalone G, Paci G (2016a) Inbreeding depression in Lizard canaries breed estimated by pedigree analysis. Czech J Anim Sci 61:15–21
Cecchi F, Paci G, Spaterna A, Ragatzu M, Ciampolini (2016b) Demographic approach on the study of genetic parameters in the dog Braque Français type Pyrénées Italian population. Ital J Anim Sci 15:1–7
Cecchi F, Carlini G, Giuliotti L, Russo C (2018) Inbreeding may affect phenotypic traits in an Italian population of Basset Hound dogs. Rend Fis Acc Lincei 29:165–170
Cecchi F, Vannucchi I, Carlini G, Macchioni F (2020) A first survey on inbreeding, phenotypic traits, coat colour and health problems prevalence in a population of English Cocker Spaniel dog. Rend Fis Acc Lincei 31:873–880
Ciampolini R, Cecchi F, Paci G, Policardo C, Spaterna A (2013) Investigation on the genetic variability of the american pit bull terrier dogs belonging to an italian breeder using microsatellite markers and genealogical data. Cytol Genet 47(4):217–221
Clark NR (2003a) The Halls Heeler. In: Clark NE (ed) Dog called Blue. WriteLight Pty Ltd, Sydney, pp 5–14
Clark NE (2003b) Kaleski’s Myths. In: Clark NE (ed) A Dog called Blue. WriteLight Pty Ltd, Sydney, pp 22–26
Cole JB, Franke DE, Leighton EA (2004) Population structure of a colony of dog guides. J Anim Sci 82:2906–2912
Corbett L (1995) The Dingo in Australia and Asia. University of New South Wales Press, Sydney
Corbett L (2001) The conservation status of the dingo Canis lupus dingo in Australia, with particular reference to New South Wales: threats to pure dingoes and potential solutions. In: Dickman CR, Lunney A (eds) Symposium on the Dingo. Royal Zoological Society of New South Wales, Mosman, pp 10–19
Dürig N, Letko A, Lepori V, Hadji-Rasouliha S, Loechel R, Kehl A, Hytönen MK, Lohi H, Mauri N, Dietrich J, Wiedmer M, Drögemüller M, Jaga nnathan V, Schmutz SM, Leeb T (2018) Two MC1R loss-of-function alleles in cream-coloured Australian Cattle Dogs and white Huskies. Anim Genet 49:284–290
DVG (2021) What is IPO / Schutzhund? http://www.dvg-america.com/testing/whatis.html Accessed by 28 Jan 2021
Elledge AE, Leung LK-P, Allen LR, Firestone K, Wilton AN (2006) Assessing the taxonomic status of dingoes Canis familiaris dingo for conservation. Mammal Rev 36:142–156
Fleming P, Corbett L, Harden R, Thomson P (2001) Managing the impacts of dingoes and other wild dogs. Bureau of Rural Sciences, Kingston
Frankham R, Ballou JD, Briscoe DA (2002) Introduction to conservation genetics. Cambridge University Press, Cambridge
Gandini GC, Bagnato A, Miglior F, Pagnacco G (1992) Inbreeding in the Italian Haflinger horse. J Anim Breed Genet 109:433–443
Gómez MD, Valera M, Molina A, Gutiérrez JP, Goyache F (2009) Assessment of inbreeding depression for body measurements in Spanish Purebred (Andalusian) horses. Livest Sci 122:149–155
Goyache F, Gutie´ rrez JP, Ferna´ ndez I, Gomez E, Alvarez I, Dı´ez J, Royo LJ. (2003) Using pedigree information to monitor genetic variability of endangered populations: the Xalda sheep breed of Asturias as an example. J Anim Breed Genet 120:95–105
Gutiérrez JP, Goyache F (2005) A note on ENDOG: a computer program for analysing pedigree information. J Anim Breed Genet 122:172–176
Harmon LJ, Braude S (2010) Conservation of small populations: Effective population size, inbreeding, and the 50/500 rule. In: Braude S, Low SB (eds) An Introduction to Methods and Models in Ecology and Conservation Biology. Princeton University Press, Princeton, pp 125–138
Kennel Working Mates (2021) About Australian Cattle Dog. https://www.workingmates.se/sv-SE/the-dogs/australian-cattle-dog-19123934 Accessed by 28 Jan 2021
Kinghorn BP (1994) Pedigree viewer—a graphical utility for browsing pedigreed data sets. In: Fifth World Congress on Genetics Applied to Livestock Production. Guelph, 7–12 August 1994. vol 22, pp 85–86
Lee P (2011) Longevity of the Australian Cattle Dog: results of a 100-dog survey. ACD Spotlight 4:96–105
Leroy G (2014) Inbreeding depression in livestock species: review and meta-analysis. Anim Genet 45:618–628
Leroy G, Rognon X, Varlet A, Joffrin C, Verrier E (2006) Genetic variability in French dog breed assessed by pedigree data. J Anim Breed Genet 123:1–9
Libro genealogico, ENCI (2021). Australian Cattledog. https://www.enci.it/libro-genealogico/razze/australian-cattledog. Accessed 1 Feb 2021
Mazzatenta M, Carluccio A, Robbe D, Di Giulio C, Cellerino A (2017) The companion dog as a unique translational model for aging. Semin Cell Dev Biol 70:141–153
Meuwissen TI, Luo Z (1992) Computing inbreeding coefficients in large populations. Genet Sel Evol 24:305–313
Ólafsdóttir GÁ, Kristjánsson T (2006) Correlated pedigree and molecular estimates of inbreeding and their ability to detect inbreeding depression in the Icelandic sheepdog, a recently bottlenecked population of domestic dogs. Conserv Genet 9:1639–1641
Pagnacco G (2004) Genetica animale applicata, 1a edn. C.E.A Casa Editrice Ambrosiana, Milano
Sabbioni A, Beretti V, Vaccari Simonini F (2008) Parametri demografici, variabilità genetica e struttura della popolazione nella razza canina Bolognese. In: Proc. LXII congr. S.I.S.Vet, San Benedetto del Tronto, Italy, pp 467–468
Sanderson A (1981) The complete book for Australian dogs. The Currawong Press Pty Ltd, Milson’s Point, pp 45–67
Sargolzaei M, Iwaisaki H, Colleau JJ (2006) CFC (Contribution, Inbreeding (F), Coancestry, Release 1.0. A software package for pedigree analysis and monitoring genetic diversity. In: Proc. 8th World Congress on Genetics Applied to Livestock Production, Belo Horizonte, Brazil (CD-ROM)
Shariflou MR, James JW, Nicholas FW, Wade CM (2011) A genealogical survey of Australian registered dog breeds. Vet J 189:203–210
Wolc A, Balińska K (2010) Inbreeding effects on exterior traits in Polish Konik horses. Arch Tierz 53:1–8
Wright S (1922) Coefficients of inbreeding and relationship. Am Nat 56:330–338
Funding
Open access funding provided by Università di Pisa within the CRUI-CARE Agreement. This work is supported by a grant from University of Pisa (Ateneo—Prof Francesca Cecchi).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors report that they have no conflicts of interest. The authors alone are responsible for the content and writing of this article.
Animal studies statement
All animal procedures used in this study were in agreement with the ethical and animal welfare concerns of the Committee on the Ethics of Animal Experiments of Minimally Invasive Surgery Centre and fully complied with recommendations outlined by the Italian laws.
Informed consent
All authors have approved this manuscript, agree to the order in which their names are listed, declare that no conflict of interest exist and deny any commercial affiliation.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Ciccarelli, J., Macchioni, F. & Cecchi, F. A genealogical survey on the main bloodline of the Australian Cattle Dog in Italy. Rend. Fis. Acc. Lincei 32, 357–364 (2021). https://doi.org/10.1007/s12210-021-00993-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12210-021-00993-3