Abstract
Introduction
AddAB enzyme is a helicase–nuclease complex that initiates recombinational repair of double-stranded DNA breaks. It catalyzes processive DNA unwinding and concomitant resection of the unwound strands, which are modulated by the recognition of a recombination hotspot called Chi in the 3′-terminated strand. Despite extensive structural, biochemical and single molecule studies, the detailed molecular mechanism of DNA unwinding by the complex and modulation by Chi sequence remains unclear.
Methods
A model of DNA unwinding by the AddAB complex and modulation by Chi recognition was presented, based on which the dynamics of AddAB complex was studied analytically.
Results
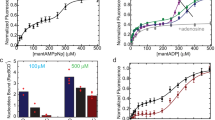
The theoretical results explain well the available experimental data on effect of DNA sequence on velocity, effect of Chi recognition on velocity, static disorder peculiar to the AddAB complex, and dynamics of pausing of wild-type and mutant AddAB complexes occurring at Chi or Chi-like sequence. Predictions were provided. Comparisons of AddAB complex with other helicase–nuclease complexes such as RecBCD and AdnAB were made.
Conclusions
The study has strong implications for the molecular mechanism of DNA unwinding by the AddAB complex. The intriguing issues are addressed of why Chi recognition is an inefficient process, how AddAB complex pauses upon recognizing Chi sequence, how the paused state transits to the translocating state, why the mutant AddAB with a stronger affinity to Chi sequence has a shorter pausing lifetime, why the pausing lifetime is sensitive to the solution temperature, and so on.








Similar content being viewed by others
Notes
It is also possible that the force arising from the cleft closing induced by ATP binding cannot overcome the strong binding force of 2A to ssDNA. Thus, it is expected that after ATP binding the cleft is still kept in the open conformation or 2A is moved in a position between that in the open cleft and that in the closed cleft by distorting the ssDNA, both causing appearance of the internal tension. These are consistent with the structural data for AddAB structures bound with DNA containing Chi sequence.10 Then, the release of ATP-hydrolysis products makes 2A in the position in the open cleft with the disappearance of the internal tension. Thus, multiple ATPase cycles take place with no movement of 1A along DNA.
References
Arnold, D. A., N. Handa, I. Kobayashi, and S. C. Kowalczykowski. A novel, 11 nucleotide variant of chi, chi*: one of a class of sequences defining the Escherichia coli recombination hotspot chi. J. Mol. Biol. 300:469–479, 2000.
Carrasco, C., N. S. Gilhooly, M. S. Dillingham, and F. Moreno-Herrero. On the mechanism of recombination hotspot scanning during double-stranded DNA break resection. Proc. Natl Acad. Sci. USA 110:E2562–E2571, 2013.
Chedin, F., N. Handa, M. S. Dillingham, and S. C. Kowalczykowski. The AddAB helicase/nuclease forms a stable complex with its cognate chi sequence during translocation. J. Biol. Chem. 281:18610–18617, 2006.
Chedin, F., and S. C. Kowalczykowski. A novel family of regulated helicases/nucleases from Gram-positive bacteria: insights into the initiation of DNA recombination. Mol. Microbiol. 43:823–834, 2002.
Dillingham, M. S., and S. C. Kowalczykowski. RecBCD enzyme and the repair of double-stranded DNA breaks. Microbiol. Mol. Biol. Rev. 72:642–671, 2008.
Gilhooly, N. S., C. Carrasco, B. Gollnick, M. Wilkinson, D. B. Wigley, F. Moreno-Herrero, and M. S. Dillingham. Chi hotspots trigger a conformational change in the helicase-like domain of AddAB to activate homologous recombination. Nucleic Acids Res. 44:2727–2741, 2016.
Glennon, T. M., J. Villa, and A. Warshel. How does GAP catalyze the GTPase reaction of Ras?: A computer simulation study. Biochemistry 39:9641–9651, 2000.
Grigorenko, B. L., A. V. Rogov, I. A. Topol, S. K. Burt, H. M. Martinez, and A. V. Nemukhin. Mechanism of the myosin catalyzed hydrolysis of ATP as rationalized by molecular modeling. Proc. Natl Acad. Sci. USA 104:7057–7061, 2007.
Kowalczykowski, S. C. Initiation of genetic recombination and recombination-dependent replication. Trends Biochem. Sci. 25:156–165, 2000.
Krajewski, W. W., X. Fu, M. Wilkinson, N. B. Cronin, M. S. Dillingham, and D. B. Wigley. Structural basis for translocation by AddAB helicase–nuclease and its arrest at chi sites. Nature 508:416–419, 2014.
Levin, M. K., M. M. Gurjar, and S. S. Patel. ATP binding modulates the nucleic acid affinity of hepatitis C virus helicase. J. Biol. Chem. 278:23311–23316, 2003.
Liu, B., R. J. Baskin, and S. C. Kowalczykowski. DNA unwinding heterogeneity by RecBCD results from static molecules able to equilibrate. Nature 500:482–485, 2013.
Saikrishnan, K., J. T. Yeeles, N. S. Gilhooly, W. W. Krajewski, M. S. Dillingham, and D. B. Wigley. Insights into Chi recognition from the structure of an AddAB-type helicase–nuclease complex. EMBO J. 31:1568–1578, 2012.
Singleton, M. R., M. S. Dillingham, M. Gaudier, S. C. Kowalczykowski, and D. B. Wigley. Crystal structure of RecBCD enzyme reveals a machine for processing DNA breaks. Nature 432:187–193, 2004.
Sinha, K. M., M. C. Unciuleac, M. S. Glickman, and S. Shuman. AdnAB: a new DSB-resecting motor–nuclease from mycobacteria. Genes Dev. 23:1423–1437, 2009.
Smith, G. R. How RecBCD enzyme and Chi promote DNA break repair and recombination: a molecular biologist’s view. Microbiol. Mol. Biol. Rev. 76:217–228, 2012.
Spies, M., I. Amitani, R. J. Baskin, and S. C. Kowalczykowski. RecBCD enzyme switches lead motor subunits in response to χ recognition. Cell 131:694–705, 2007.
Spies, M., P. R. Bianco, M. S. Dillingham, N. Handa, R. J. Baskin, and S. C. Kowalczykowski. A molecular throttle: The recombination hotspot χ controls DNA translocation by the RecBCD helicase. Cell 114:647–654, 2003.
Unciuleac, M. C., and S. Shuman. Double strand break unwinding and resection by the mycobacterial helicase–nuclease AdnAB in the presence of single strand DNA-binding protein (SSB). J. Biol. Chem. 285:34319–34329, 2010.
Unciuleac, M. C., and S. Shuman. Characterization of the mycobacterial AdnAB DNA motor provides insights into the evolution of bacterial motor–nuclease machines. J. Biol. Chem. 285:2632–2641, 2010.
Wigley, D. B. Bacterial DNA repair: recent insights into the mechanism of RecBCD, AddAB and AdnAB. Nat. Rev. Microbiol. 11:9–13, 2013.
Wilkinson, M., Y. Chaban, and D. B. Wigley. Mechanism for nuclease regulation in RecBCD. eLife 5:e18227, 2016.
Wu, P., S. Nakano, and N. Sugimoto. Temperature dependence of thermodynamic properties for DNA/DNA and RNA/DNA duplex formation. Eur. J. Biochem. 269:2821–2830, 2002.
Xie, P. Dynamics of monomeric and hexameric helicases. Biophys. Chem. 211:49–58, 2016.
Xie, P. Processivity of nucleic acid unwinding and translocation by helicases. Proteins 84:1590–1605, 2016.
Xie, P. Dynamics of DNA unwinding by helicases with frequent backward steps. Math. Biosci. 294:33–45, 2017.
Xie, P. A model of DNA unwinding dynamics by the RecBCD complex and its regulation by Chi recognition. J. Theor. Biol. 448:142–156, 2018.
Yeeles, J. T., and M. S. Dillingham. The processing of double-stranded DNA breaks for recombinational repair by helicase–nuclease complexes. DNA Repair 9:276–285, 2010.
Yeeles, J. T., K. van Aelst, M. S. Dillingham, and F. Moreno-Herrero. Recombination hotspots and single-stranded DNA binding proteins couple DNA translocation to DNA unwinding by the AddAB helicase–nuclease. Mol. Cell 42:806–816, 2011.
Acknowledgment
This work was supported by the National Natural Science Foundation of China (Grant No. 11775301).
Conflict of interests
Ping Xie declares that he has no conflicts of interest.
Ethical standards
No human studies were carried out by the author for this article. No animal studies were carried out by the author for this article.
Author information
Authors and Affiliations
Corresponding author
Additional information
Associate Editor Edward Sander oversaw the review of this article.
Rights and permissions
About this article
Cite this article
Xie, P. Modeling DNA Unwinding by AddAB Helicase–Nuclease and Modulation by Chi Sequences: Comparison with AdnAB and RecBCD. Cel. Mol. Bioeng. 12, 179–191 (2019). https://doi.org/10.1007/s12195-018-00563-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12195-018-00563-y