Abstract
Vaccinations are widely credited with reducing death rates from COVID-19, but the underlying host-viral mechanisms/interactions for morbidity and mortality of SARS-CoV-2 infection remain poorly understood. Acute respiratory distress syndrome (ARDS) describes the severe lung injury, which is pathologically associated with alveolar damage, inflammation, non-cardiogenic edema, and hyaline membrane formation. Because proteostatic pathways play central roles in cellular protection, immune modulation, protein degradation, and tissue repair, we examined the pathological features for the unfolded protein response (UPR) using the surrogate biomarker glucose-regulated protein 78 (GRP78) and co-receptor for SARS-CoV-2. At autopsy, immunostaining of COVID-19 lungs showed highly elevated expression of GRP78 in both pneumocytes and macrophages compared with that of non-COVID control lungs. GRP78 expression was detected in both SARS-CoV-2-infected and un-infected pneumocytes as determined by multiplexed immunostaining for nucleocapsid protein. In macrophages, immunohistochemical staining for GRP78 from deceased COVID-19 patients was increased but overlapped with GRP78 expression taken from surgical resections of non-COVID-19 controls. In contrast, the robust in situ GRP78 immunostaining of pneumocytes from COVID-19 autopsies exhibited no overlap and was independent of age, race/ethnicity, and gender compared with that from non-COVID-19 controls. Our findings bring new insights for stress-response pathways involving the proteostatic network implicated for host resilience and suggest that targeting of GRP78 expression with existing therapeutics might afford an alternative therapeutic strategy to modulate host-viral interactions during SARS-CoV-2 infections.
Similar content being viewed by others
Introduction
The coronavirus disease-19 (COVID-19) pandemic has afflicted over 33 million people, causing approximately 5 million hospitalizations and more than 600,000 deaths in the USA alone (> 4 million deaths worldwide). Understanding mechanisms through which the severe acute respiratory disease coronavirus 2 (SARS-CoV-2) infection promotes major pathophysiological consequences of COVID-19 is key to improving strategies for monitoring, mitigation, and/or treatment. Host stress-response pathways orchestrate a cascade of cellular events as the first line of defense to preserve cellular homeostasis by maintaining proper protein folding, i.e., “proteostasis” (Benjamin & McMillan, 1998; Christians & Benjamin, 2012). For example, the endoplasmic reticulum (ER)-localized glucose-regulated protein 78 (GRP78/HSPA5) or binding immunoglobulin protein (BiP), a molecular chaperone and mediator of the unfolded protein response (UPR), plays a key role involving the proteostatic network/machinery during ER stress (Balch et al., 2014; Pehote & Vij, 2020) and has also has been implicated in host-viral interactions. While the spike protein of SARS-CoV-2 binds to the ACE2 receptor in humans, host tropism is also determined by GRP78/HSPA5, which acts as a co-receptor on the cell surface and promotes viral entry for the SARS-CoV, Middle East respiratory syndrome coronavirus (MERS-CoV), bat coronavirus (Chu et al., 2018), and has been predicted to bind SARS-CoV-2 spike protein/ACE2 complexes (Elfiky, 2021; Ha et al., 2020; Ibrahim et al., 2020). Because viral genomes lack heat shock proteins (HSPs), invasion of the single-stranded RNA SARS-CoV-2 virus is obligatorily coupled with, and dependent upon, recruitment of the host’s protein synthesis machinery to maintain folding of viral proteins. Competition for the host’s HSP/GRP78 machinery creates existential threats for both host and virus, invoking complex cell fate decisions resulting in either death or survival. Recent evidence for the appearance of plasma UPR biomarkers with COVID-19 suggests early tissue damage associated with the hyperinflammatory state. However, insight about clinical progression to severe disease (Koseler et al., 2020), critical for timely management of COVID-19 patients (Aguiar et al., 2020), is lacking. We hypothesized that host-viral interactions of SARS-CoV-2 are conceivably accompanied by defects either in the components of the proteostasis machinery, in the dysregulation of normal proteostatic signaling, or both. GRP78/HSPA5 is expressed in the normal respiratory airway epithelial cells and to a lesser extent in normal alveolar pneumocytes, and can facilitate pathogenic viral entry into the cells (Ibrahim et al., 2019). Elevated levels of circulating grp78 mRNA have been reported in COVID-19 patients (Ibrahim et al., 2019). We therefore focused on the expression of GRP78 in lungs of patients suffering COVID-19 deaths with respiratory failure.
Materials and methods
Materials
A monoclonal rabbit antibody to GRP78 (BiP (C50B12) Rabbit mAb; Cell Signaling, Danvers, MA; Cat#3177; 1:200) and Envision Plus polymer (Agilent/DAKO) were used with magenta chromogen for visualization. Immunohistochemistry for SARS-CoV-2 nucleocapsid protein was performed using an affinity-purified polyclonal rabbit antibody (affinity-purified rabbit IgG; ProSci, Poway, CA; Cat# 9099; 0.04 µg/ml). Multiplex immunofluorescence staining of GRP78 was visualized using the TSA Plus Cyanine 3 System, Perkin Elmer, Waltham, MA: Cat# NEL744B001KT. SARS-CoV-2 nucleocapsid was visualized using the TSA Plus Cyanine 5 System (Perkin Elmer Cat# NEL745001KT). Macrophages/monocytes were identified using CD68 (CD68 Monoclonal Antibody KP1, Thermo Fisher Scientific, Grand Island, NY; Cat# 14-0688-82) and visualized using the TSA Plus Cyanine 7 System (Perkin Elmer, Cat# CUSM03644000EA) following the addition of peroxidase blocking reagent (hydrogen peroxide solution, Sigma-Aldrich, St. Louis, MO; Cat# H1009). Antibody stripping was accomplished using the Omnis (Dako/Agilent) low (CD68 and SARS-CoV-2 nucleocapsid) or high (GRP78) pH antigen retrieval protocol. Pneumocytes were identified using anti-pan-cytokeratin antibody (mouse monoclonal AE1/AE3 blend, Cat# M3515, Agilent) followed by Alexa-Fluor-488-labeled secondary antibody (Cat# A11029, Thermo Fisher Scientific).
Immunohistochemistry
Immunohistochemistry was performed on deparaffinized sections of lungs either from COVID-19 decedents or from patients with non-COVID-19 conditions using an Omnis autostainer (Agilent/DAKO, Santa Clara, CA). A monoclonal rabbit antibody to GRP78 (BiP; C50B12; Cell Signaling, Danvers, MA; Cat# 3177; 1:200 dilution) and Envision Plus polymer (Agilent/DAKO) were used with magenta chromogen for bright-field visualization. For fluorescence immunohistochemistry, SARS-CoV-2 nucleocapsid protein was examined using an affinity-purified polyclonal rabbit antibody (ProSci, Poway, CA; Cat# 9099) in a cross-validated protocol as previously described (Sun, 2020). Multiplex immunofluorescence staining of GRP78, SARS-CoV-2 nucleocapsid, and CD68 was performed following incubation with the peroxidase blocking reagent (hydrogen peroxide solution, Sigma-Aldrich, St. Louis, MO; Cat# H1009) using iterative incubation with (a) primary antibody, (b) secondary HRP-conjugated antibody, (c) HRP-mediated deposition of fluorescent-tyramide, and (d) antibody stripping using the Omnis low pH (pH 6; CD68 and SARS-CoV-2 nucleocapsid) or high pH (pH 9; GRP78) antigen retrieval protocols, repeated for each of the three primary antibodies. Specifically, immunofluorescence staining of GRP78 (C50B12; Cell Signaling, Danvers, MA; Cat#3177; 1:800 dilution for IF) was visualized using the TSA Plus Cyanine-3 System (Perkin Elmer, Waltham, MA; Cat# NEL744B001KT); SARS-CoV-2 nucleocapsid (ProSci, Poway, CA; Cat# 9099; 0.01 µg/ml) was visualized using the TSA Plus Cyanine-5 System (Perkin Elmer Cat# NEL745001KT); macrophages were identified using CD68 (CD68 Monoclonal Antibody KP1, Thermo Fisher Scientific, Grand Island, NY; Cat# 14-0688-82; 1:10,000 dilution) and visualized using the TSA Plus Cyanine-7 System (Perkin Elmer, Cat# CUSM03644000EA). Iterative primary antibody stripping was accomplished using the Omnis (Dako/Agilent) low (pH 6; CD68 and SARS-CoV-2 nucleocapsid) or high (pH 9; GRP78) antigen retrieval protocols. Pneumocytes were identified using anti-pan-cytokeratin antibody (mouse monoclonal AE1/AE3 blend, Cat# M3515, Agilent) followed by Alexa-Fluor-488-labeled secondary antibody (Cat# A11029, Thermo Fisher Scientific) and final counterstaining with DAPI. After coverslipping, chromogen IHC–stained slides were scanned using the bright-field mode of Pannoramic 250 (3DHistech Ltd., Budapest, Hungary), and images were generated using Caseviewer, while multicolor immunofluorescence–stained slides were scanned on a Vectra Polaris imaging system (Akoya Biosciences) and images generated from PhenoCharts (Akoya Biosciences, Marlborough, MA).
Lung sections analyzed were taken from the most prominent areas of change on gross examination as determined by a pathologist from the COVID-19-positive group. Sections from non-COVID-19 control specimens were selected from grossly unremarkable lung tissue. Each lung section of every case was evaluated microscopically in its entirety at low and high magnifications by pathologists. Representative images showing the dominant histological features were taken at different magnifications (20 × , 200 × , and 400 ×) from each case in both the COVID-19 and the non-COVID-19 control groups for scoring.
Scoring of GRP78 expression
GRP78 expression was evaluated in pneumocytes and macrophages by pathologists. The GRP78 staining intensity of either cell type was awarded the following 4 grades according to the staining intensity of magenta color as follows: 0—no staining, 1—weak, 2—moderate, and 3—strong. The proportion of either pneumocytes or macrophages of each tissue that were positive for GRP78 was also graded as follows: 1—less than 5%; 2—5-24%, 3—25-49%, 4—50-74%, and 5—75-100%. The sum of the GRP78 intensity grade and positivity grade for each cell type generated an overall GRP78 score that was represented by a 4-step scale as follows: grades 1–2—negative ( −); grades 3-4—weak positive ( +); grades 5-6—moderate positive (+ +); and grades 7-8—strong positive (+ + +).
Statistical methods
Continuous variables are summarized as median (minimum, maximum) and groups are compared using exact Mann–Whitney or Kruskal–Wallis tests. Categorical variables are summarized as number (%) and compared using exact Fisher’s tests. p values are unadjusted, and any p < 0.05 is denoted as significant. The software used was the Statistical Package for Social Sciences v26.
Results
Autopsy tissues from 12 patients diagnosed with COVID-19 (based on PCR tests and consistent CT chest, lab tests, and clinical exam) who were admitted through the emergency department of the Froedtert & the Medical College of Wisconsin regional hospital and died from SARS-CoV-2 pneumonia were included in the study. Archival non-COVID-19 control lung tissues were from resections (n = 25; 14 cases of malignancy; samples secured from the lung remote from that involved with tumor; and 11 cases showing inflammation/infection). For the control lung tissues with remote neoplasm, lungs were resected most commonly for either adenocarcinoma (n = 8) or squamous cell carcinoma (n = 4). For control lung tissues without malignancy, inflammation either with or without granulomatous changes was the most frequent pathological diagnosis (n = 11), followed by either interstitial lung disease or fibrosis (n = 4). Pathological evidence of emphysema was evident in tissue of 3 cases with cancer and 4 cases with inflammatory changes. All samples from COVID-19 autopsy patients demonstrated diffuse alveolar damage throughout the lungs.
Demographic information of the patient populations is shown in Table 1. Age and gender distribution of COVID-19 patients was similar to that of controls, including those with either cancer or inflammatory diseases. Of the COVID-19 patients, 67% were female, and 45% and 57% of the non-COVID-19/no cancer cases and non-COVID-19/remote cancer were females, respectively (p = 0.58). Median (range) ages were 69 (28–91) for COVID-19 cases; 60 (22–70) for non-COVID-19/no cancer cases; and 62.5 (56–85) for non-COVID-19/remote cancer cases (p = 0.19). The proportion of Black cases was higher among COVID-19 cohort (7 Blacks, 5 Whites) compared to that among non-COVID-19 controls (1 Blacks, 11 Whites; p < 0.001). These data are consistent with the reported greater burden of COVID-19 in Black Americans (Price-Haywood et al., 2020).
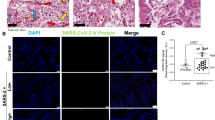
To determine the status of GRP78 protein expression in lungs of COVID-19 patients, we performed immunohistochemistry of GRP78. Figure 1 shows representative images of lungs from COVID-19 and non-COVID-19 patients stained for GRP78 protein expression by immunohistochemistry. Both intensity and distribution for GRP78 expression (magenta staining) were easily visible and more abundant in samples from COVID-19 patients compared with those from control patients. Parallel multiplexed co-immunostaining verified abundant GRP78 expression in a large proportion of both CD68+ macrophages and pan-cytokeratin-positive pneumocytes in lung tissue from a patient who died during the active viral infection phase (Fig. 2a, b). GRP78 was also expressed within actively infected pneumocytes, as visualized by the co-expression of SARS-CoV-2 nucleocapsid protein (Fig. 2c–e).
GRP78 protein expression among the lung sections from patients with COVID-19 (upper panels) and non-COVID-19 controls (bottom panels; a, b, g, h at 20 × ; c, d, i, k, l at 200 × ; and e, f at 400 ×). Yellow arrows indicate pneumocytes and green arrows identify macrophages. (a, b) Representative overview histology images showing intense GRP78 expression in COVID-19 lung; (c–f) Moderate to strong positive pneumocytes and macrophages on higher magnification in COVID-19 lung; (g, h) overview histology showing lung from non-COVID-19 control group; (i–l) rare positive pneumocytes and weak to moderate positive macrophages on higher magnification in lung from non-COVID-19 controls
Multiplexed immunostaining of COVID-19 lung showing GRP78 expression in macrophages and pneumocytes. (a) Overview histology showing pneumocytes (pan-cytokeratin; brown) and macrophages (CD68; green) with nuclei visualized by DAPI staining (blue). (b) Higher power frame from (a) with pneumocytes (brown) and macrophages (green) and merged with GRP78 staining (purple); (c) the same frame with pneumocytes (brown) and macrophages (green) and merged with SARS-CoV-2 nucleocapsid staining (yellow); (d) the same frame with GRP78 (purple) merged with SARS-CoV-2 nucleocapsid staining (yellow); and (e) all five colors merged
Table 2 shows the distribution of GRP78 staining intensity grades and percent GRP78 positivity grades for pneumocytes and macrophages on a case-by-case basis, and the two grades were added to yield a composite intensity plus positivity GRP78 score for each of the two cell types. Moderate to high GRP78 staining scores in the pneumocytes in lungs from COVID-19 patients were markedly higher than those in the pneumocytes from non-COVID-19 control lungs (p < 0.001). Similarly, strong positive scores of GRP78 in the macrophages were higher in the COVID-19 group compared with those in the non-COVID-19 controls (p < 0.001). Interestingly, the higher score for GRP78 expression in macrophages from COVID-19 patients was attributable to higher intensity of staining rather than the percentage of GRP78-positive cells. The subgroup of non-COVID-19 control with resections for cancer exhibited similar differences between pneumocyte scores relative to that of COVID-19 patients (COVID-19: median 8 (n = 6–8); cancer median 3 (n = 24); p < 0.001). When the analysis excluded data from Black cases, similar differences in scoring for pneumocyte and macrophage GRP78 expression between COVID-19 and control cases persisted (14.6 and 11.5 for COVID-19 patients respectively; p < 0.001).
Figure 3 shows the distribution of GRP78 scores across all age ranges and stratified by COVID-19 versus non-COVID-19 controls. In pneumocytes (Fig. 3a), there was a clear separation of GRP78 scores across all age groups with no overlap of patients with COVID-19 and other groups. Similarly, the same trend in the separation can be seen for GRP78 immunostaining in macrophages (Fig. 3b), but unlike the data from pneumocytes, there was a partial overlap in GRP78 scores in COVID-19 and non-COVID-19 controls.
a shows the scoring of pneumocytes for GRP78 staining as a function of diagnosis (COVID-19 or control states) and stratified by age. There is a clear separation of scores for GRP78 expression that is sustained across all age ranges studied. b shows the same data for macrophages. Scores for COVID-19 are greater than those for non-COVID-19 controls, but there is an overlap between the scores for the group, unlike that of data for pneumocytes
Discussion
Our analyses show substantially increased expression of the stress protein GRP78 and SARS-CoV-2 co-receptor in pneumocytes and lung macrophages of patients who died while hospitalized for COVID-19 relative to those of non-COVID-19 control lungs. These observations may provide insights into the source of circulating GRP78 mRNA and protein in patients with SARS-COV-2-positive pneumonia that has been observed relative to individuals with SARS-COV-2-negative pneumonia or a healthy group (Palmeira et al., 2020). While GRP78 serves key cytoprotective and homeostatic effects in cells under some conditions, including viral infection and associated inflammatory states, GRP78 is also considered a co-receptor with angiotensin-converting enzyme 2 (ACE2) for SARS-CoV-2. Our observation in COVID-19 pneumonia lungs, that elevated GRP78 expression levels were detected within both SARS-CoV-2 infected (nucleocapsid protein–positive) and un-infected (nucleocapsid protein–negative) pneumocytes determined by multiplexed immunostaining, raises the possibility of a SARS-CoV-2 virus-GRP78 feed-forward loop that facilitates disease progression. In support of this notion, a humanized monoclonal antibody to GRP78 was recently shown to not only reduce cell surface ACE2 but to decrease SARS-CoV-2 entry and infection (Carlos et al., 2021). ACE2 expression is reported to increase with age in patients requiring mechanical ventilation (Baker et al., 2021), raising the additional possibility that mechanical ventilation may contribute to the observed increased GRP78 levels in pneumocytes via enhanced ACE2 in the autopsied COVID-19 lungs. However, age-related increase in ACE2 expression from mechanical ventilation alone cannot account for our observations because increased GRP78 scores in pneumocytes and macrophages were observed across all age groups with COVID-19, without a relationship to age. Moreover, four of the 12 COVID-19 patients analyzed were not treated with mechanical ventilation; and the median GRP78 scores for pneumocytes and macrophages were not different from those who did receive mechanical ventilation (8 vs. 8 for pneumocytes; 7 vs. 8 for macrophages). Further work will be required to determine the possible direct and/or indirect effects of SARS-CoV-2 infection on the upregulation of GRP78 in pneumocytes. Our observations provide new insights into stress-response pathways involving the proteostatic network implicated for host resilience and support the notion that targeting of GRP78 early in the disease process may disrupt an accelerating disease progression and might represent an alternative therapeutic strategy.
What is GRP78?
The HSPA5 gene encodes the 78-kilo-Dalton glucose-regulated protein (GRP78), which is also known as BiP and acts as a central regulator of ER homeostasis and whose upregulation is widely used as a sentinel marker for ER stress under pathologic conditions (Ni & Lee, 2007; Pobre et al., 2019). The unfolded protein response (UPR) includes transcriptional activation of IRE1α (inositol-requiring enzyme 1α), PERK (pancreatic endoplasmic reticulum kinase), ATF6 (activating transcription factor 6), and X-box binding protein 1 (XBP1) to facilitate increases of UPR target genes encoding chaperone protein GRP78 in order to restore ER homeostasis (Ogata et al., 2019; Yoshida et al., 2001). In addition to the ER, GRP78 protein has been reported to have other compartment-specific locations such as the cell surface, the mitochondrion, and nucleus. Chaperone GRP78 expression is recognized diffusely in normal lung cells, such as bronchial surface epithelial cells, chondrocytes of the bronchial cartilage, serous cells of the bronchial glands, and alveolar macrophages (Ogata et al., 2019). Our observations support elevated GRP78 expression in pneumocytes and alveolar macrophages (Fig. 1).
Unfolded protein response in host homeostasis
In response to stressful conditions (infection, chemicals, ischemia, etc.), cells activate genetic programs which increase the synthesis of evolutionarily conserved stress proteins (heat shock proteins and GRPs) as the first line of defense to restore cellular homeostasis by maintaining proper protein folding. Competition for the host’s HSP/GRP machinery creates existential threats for both host and virus, invoking complex cell fate decisions resulting in death or survival. Viral infection induces ER stress and upregulation of the unfolded protein response (UPR); furthermore, the increased synthesis and translocation of multifunctional GRP78 to the cell surface has been implicated in enhanced infection in a positive feedback loop (Ha et al., 2020; Tsai et al., 2015). Major perturbations of subsequent proteostatic stress have been stymied by inadequate progress on the development of clinical tools to monitor proteostasis in vitro and in vivo especially in humans. Our findings raise the intriguing suggestion that GRP78 expression during SARS-CoV-2 infections could be an excellent target to track proteostatic dysregulation. In fact, several therapeutics inhibit GRP78 (Tao et al., 2019) and/or may have efficacy in SARS-CoV-2 infections via modifying GRP78 activity (Dyall et al., 2014; Palmeira et al., 2020; Zhang et al., 2021).
Localization of GRP78 in COVID-19 lungs
Whereas SARS-CoV-2 exploits the attachment and cell fusion via the ACE2 receptor, emerging evidence implicates other host cofactors for viral entry and disease pathogenesis (Cantuti-Castelvetri et al., 2020; Zamorano Cuervo & Grandvaux, 2020). Molecular modeling, docking, and structural bioinformatics that have postulated an association between molecular chaperone GRP78 and the receptor binding domain (RBD) of the spike protein of SARS-CoV-2 have given rise to the hypothesis that GRP78 facilitates virus attachment and host cell entry (Elfiky & Ibrahim, 2021; Koseler et al., 2020). GRP78 has been recently implicated in trafficking, localizing, and affording for a mechanism of dynamically regulating ACE2 expression in human lung or other receptors for SARS-CoV-2 on the cell surface (Carlos et al., 2021). In addition to TMPRSS2, GRP78 protein is found in vitro in airway epithelial cells and confirm broad in situ protein expression of GRP78 in the respiratory mucosa (Aguiar et al., 2020). These observations and suggestions are of particular interest considering the increased expression of GRP78 we observed in the lungs of patients who died of COVID-19 (Fig. 1).
GRP78 as a biomarker of disease severity
Significantly higher serum GRP78 protein and GRP78 mRNA levels in patients with SARS-CoV-2-positive pneumonia are described compared with individuals with SARS-COV-2-negative pneumonia and healthy group (Palmeira et al., 2020); however, there was no difference between SARS-CoV-2-positive pneumonia and CT-negative COVID-19 infection group (Sabirli et al., 2021). A recent proteomic analysis of 144 autopsy samples from seven organs in 19 COVID-19 patients demonstrated that of 11,394 proteins in these samples, nearly half were perturbed in the COVID-19 patients compared with those in controls. Cathepsin L1, rather than ACE2, was upregulated in the lung from the COVID-19 patients (Nie et al., 2021). Not surprisingly, systemic hyperinflammation and dysregulation of glucose and fatty acid metabolism were detected in multiple organs (Nie et al., 2021). Our report adds GRP78 to the list of proteins upregulated in lung tissue of infected patients.
Implications of ER stress and increase GRP78 expression in COVID-19 lungs
Recent reports describe markedly elevated levels of cell-free DNA (cfDNA) in the serum of patients with COVID-19, the sources of which include vascular endothelial cells and lungs (Andargie et al., 2021), and may be markers of the severity of illness. Alveolar epithelial and endothelial cells exhibit expression of hyper-phosphorylated STAT3 and immune checkpoint molecules (PD-L1 and IDO) (Chilosi et al., 2021), cells which we observed to express GRP78. Exosome formation is significantly enhanced under ER stress in humans and rodents (Kanemoto et al., 2016; Mastronardi et al., 2011). There are some reports that exosomes may have protective functions in sepsis. Circulating platelet and endothelial cell microparticles from patients with septic shock exert a protective role against vascular hypo-reactivity in rodents (Mostefai et al., 2008). There is also evidence that circulating exosomes contribute to inflammatory lung injury as well as pulmonary activation or expression of GRP78. Microparticles from septic humans increased the expression of endothelial and inducible nitric oxide synthases, cyclooxygenase-2, and nuclear factor-kβ in murine tissues. Increased levels of plasma tumor necrosis factor (TNF)-a, interleukin (IL)-6, total cell counts, polymorphonuclear (PMN) leukocyte differential cell percentages, and myeloperoxidase (MPO) activity in bronchoalveolar lavage fluid (BALF), and increased wet/dry lung weight ratios and protein concentrations in BALF are reported in mice after exosome injection but not in mice treated with exosome-depleted serum (Tang et al., 2020). A marked increase in BiP/GRP78 expression was observed in lung homogenates of mice treated with lipopolysaccharides (LPS) (Leonard et al., 2019). Mice that were genetically defective in a holoenzyme that cleaves and inactivates BiP/GRP78 were protected from LPS-induced lung injury. These data support upregulated expression and proinflammatory roles for sepsis-induced pulmonary intracellular (and not cell surface) GRP78. These reports do not identify either the cellular sources or mediators of exosomal constituents that may modify pulmonary inflammation or expression of GRP78. They are, however, consistent with our observations of enhanced expression of GRP78 in COVID-19 autopsy studies. Our studies do not permit us to address the function of increased GRP78 expression in lungs, but they provide vital new information upon which to build upon a more complete understanding of this pathway in SARS-CoV-2 pneumonia.
Limitations to our report
First, we are cautious not to overinterpret that increased levels of viral-induced synthesis of multifunctional GRP78 in COVID-19 decedents are causally related to lung injury or a recovery response. In this study, we selected tissue samples of survivors with lung resections for GRP78 expression as controls (Table 1). Our results show substantially elevated levels of GRP78 expression in pneumocytes and lung macrophages from patients with COVID-19 relative to those of persons with other disease processes including cancer. While small numbers of patients in all groups limit conclusions based on segmented data, the ages of the COVID-19 and non-COVID-19 patients are similar. GRP78 expression is known to be increased in malignancies (Wu et al., 2020), but our data show low GRP78 expression in pneumocytes and macrophages in tumor-adjacent “normal” lung tissue. There are differences in racial distribution in COVID-19 versus that in non-COVID-19 control groups with more Blacks than Whites in the COVID-19 group than that in control samples; therefore, we cannot exclude the possibility that race may modify pulmonary GRP78 expression. However, when we limited our analyses to Whites in the COVID-19 and non-COVID-19 groups, the differences in pneumocyte and macrophage GRP78 scores remained unaffected. Our semi-quantitative analyses on increased GRP78 expression of COVID-19 autopsies do not preclude dysregulation and, perhaps, proteostasis collapse per se as has been reported for other cellular processes such as glucose and fatty acid metabolism, immune, angiogenesis, and coagulation pathways (Caramaschi et al., 2021; Nie et al., 2021). Plasma levels of GRP78 are not available for our patients. Nevertheless, higher serum GRP78 protein and GRP78 mRNA levels in patients with SARS-COV-2-positive pneumonia have already been described relative to a group of individuals with SARS-COV-2-negative pneumonia or a healthy group (Palmeira et al., 2020).
Data availability
Consistent with National Institute of Health (NIH) plans for resource sharing, academic investigators will be granted access to deidentified information upon either publication and/or completion of our published and ongoing studies.
References
Aguiar JA, Tremblay BJ, Mansfield MJ, Woody O, Lobb B, Banerjee A, ... Hirota JA (2020) Gene expression and in situ protein profiling of candidate SARS-CoV-2 receptors in human airway epithelial cells and lung tissue. Eur Respir J 56(3). https://doi.org/10.1183/13993003.01123-2020
Andargie TE, Tsuji N, Seifuddin F, Jang MK, Yuen PS, Kong H, ..., Agbor-Enoh S (2021) Cell-free DNA maps COVID-19 tissue injury and risk of death and can cause tissue injury. JCI Insight, 6(7). https://doi.org/10.1172/jci.insight.147610
Baker SA, Kwok S, Berry GJ, Montine TJ (2021) Angiotensin-converting enzyme 2 (ACE2) expression increases with age in patients requiring mechanical ventilation. PLoS ONE 16(2):e0247060. https://doi.org/10.1371/journal.pone.0247060
Balch WE, Sznajder JI, Budinger S, Finley D, Laposky AD, Cuervo A M, ..., Gan W (2014) Malfolded protein structure and proteostasis in lung diseases. Am J Respir Crit Care Med 189(1) 96-103. https://doi.org/10.1164/rccm.201306-1164WS
Benjamin IJ, McMillan DR (1998) Stress (heat shock) proteins: molecular chaperones in cardiovascular biology and disease. Circ Res 83(2):117–132. https://doi.org/10.1161/01.res.83.2.117
Cantuti-Castelvetri L, Ojha R, Pedro LD, Djannatian M, Franz J, Kuivanen S, ..., Simons M (2020) Neuropilin-1 facilitates SARS-CoV-2 cell entry and infectivity. Science 370(6518) 856-860. https://doi.org/10.1126/science.abd2985
Caramaschi S, Kapp ME, Miller SE, Eisenberg R, Johnson J, Epperly G, ..., Giannico GA (2021) Histopathological findings and clinicopathologic correlation in COVID-19: a systematic review. Mod Pathol. https://doi.org/10.1038/s41379-021-00814-w
Carlos AJ, Ha DP, Yeh DW, Van Krieken R, Tseng CC, Zhang P, ..., Lee AS (2021) The chaperone GRP78 is a host auxiliary factor for SARS-CoV-2 and GRP78 depleting antibody blocks viral entry and infection. J Biol Chem 100759. https://doi.org/10.1016/j.jbc.2021.100759
Chilosi M, Poletti V, Ravaglia C, Rossi G, Dubini A, Piciucchi S, ..., Doglioni C (2021) The pathogenic role of epithelial and endothelial cells in early-phase COVID-19 pneumonia: victims and partners in crime. Mod Pathol. https://doi.org/10.1038/s41379-021-00808-8
Christians ES, Benjamin IJ (2012) Proteostasis and REDOX state in the heart. Am J Physiol Heart Circ Physiol 302(1):H24-37. https://doi.org/10.1152/ajpheart.00903.2011
Chu H, Chan CM, Zhang X, Wang Y, Yuan S, Zhou J, ..., Yuen KY (2018) Middle East respiratory syndrome coronavirus and bat coronavirus HKU9 both can utilize GRP78 for attachment onto host cells. J Biol Chem 293(30) 11709-11726. https://doi.org/10.1074/jbc.RA118.001897
Dyall J, Coleman CM, Hart BJ, Venkataraman T, Holbrook MR, Kindrachuk J, ..., Frieman MB (2014) Repurposing of clinically developed drugs for treatment of Middle East respiratory syndrome coronavirus infection. Antimicrob Agents Chemother 58(8) 4885-4893. https://doi.org/10.1128/AAC.03036-14
Elfiky AA (2021) Natural products may interfere with SARS-CoV-2 attachment to the host cell. J Biomol Struct Dyn 39(9):3194–3203. https://doi.org/10.1080/07391102.2020.1761881
Elfiky AA, Ibrahim IM (2021) Host-cell recognition through GRP78 is enhanced in the new UK variant of SARS-CoV-2, in silico. J Infect 82(5):186–230. https://doi.org/10.1016/j.jinf.2021.01.015
Ha DP, Van Krieken R, Carlos AJ, Lee AS (2020) The stress-inducible molecular chaperone GRP78 as potential therapeutic target for coronavirus infection. J Infect 81(3):452–482. https://doi.org/10.1016/j.jinf.2020.06.017
Ibrahim IM, Abdelmalek DH, Elfiky AA (2019) GRP78: A cell’s response to stress. Life Sci 226:156–163. https://doi.org/10.1016/j.lfs.2019.04.022
Ibrahim IM, Abdelmalek DH, Elshahat ME, Elfiky AA (2020) COVID-19 spike-host cell receptor GRP78 binding site prediction. J Infect 80(5):554–562. https://doi.org/10.1016/j.jinf.2020.02.026
Kanemoto S, Nitani R, Murakami T, Kaneko M, Asada R, Matsuhisa K, ..., Imaizumi K (2016) Multivesicular body formation enhancement and exosome release during endoplasmic reticulum stress. Biochem Biophys Res Commun 480(2) 166-172. https://doi.org/10.1016/j.bbrc.2016.10.019
Koseler A, Sabirli R, Goren T, Turkcuer I, Kurt O (2020) Endoplasmic reticulum stress markers in SARS-COV-2 infection and pneumonia: case-control study. Vivo 34(3 Suppl):1645–1650. https://doi.org/10.21873/invivo.11956
Leonard A, Grose V, Paton AW, Paton JC, Yule DI, Rahman A, Fazal F (2019) Selective inactivation of intracellular BiP/GRP78 attenuates endothelial inflammation and permeability in acute lung injury. Sci Rep 9(1):2096. https://doi.org/10.1038/s41598-018-38312-w
Mastronardi ML, Mostefai HA, Meziani F, Martinez MC, Asfar P, Andriantsitohaina R (2011) Circulating microparticles from septic shock patients exert differential tissue expression of enzymes related to inflammation and oxidative stress. Crit Care Med 39(7):1739–1748. https://doi.org/10.1097/CCM.0b013e3182190b4b
Mostefai HA, Meziani F, Mastronardi ML, Agouni A, Heymes C, Sargentini C, ..., Andriantsitohaina R (2008) Circulating microparticles from patients with septic shock exert protective role in vascular function. Am J Respir Crit Care Med 178(11) 1148-1155. https://doi.org/10.1164/rccm.200712-1835OC
Ni M, Lee AS (2007) ER chaperones in mammalian development and human diseases. FEBS Lett 581(19):3641–3651. https://doi.org/10.1016/j.febslet.2007.04.045
Nie X, Qian L, Sun R, Huang B, Dong X, Xiao Q, ..., Guo T (2021) Multi-organ proteomic landscape of COVID-19 autopsies. Cell 184(3) 775–791 e714. https://doi.org/10.1016/j.cell.2021.01.004
Ogata S, Kameda K, Kono T, Ozeki Y, Hashimoto H, Tominaga S, Nakanishi K (2019) Expressions of ATF6, XBP1, and GRP78 in normal tissue, atypical adenomatous hyperplasia, and adenocarcinoma of the lung. Hum Pathol 83:22–28. https://doi.org/10.1016/j.humpath.2018.08.009
Palmeira A, Sousa E, Koseler A, Sabirli R, Goren T, Turkcuer I, ..., Vasconcelos MH (2020) Preliminary virtual screening studies to identify GRP78 inhibitors which may interfere with SARS-CoV-2 infection. Pharmaceuticals (Basel) 13(6). https://doi.org/10.3390/ph13060132
Pehote G, Vij N (2020) Autophagy augmentation to alleviate immune response dysfunction, and resolve respiratory and COVID-19 exacerbations. Cells 9(9). https://doi.org/10.3390/cells9091952
Pobre KFR, Poet GJ, Hendershot LM (2019) The endoplasmic reticulum (ER) chaperone BiP is a master regulator of ER functions: getting by with a little help from ERdj friends. J Biol Chem 294(6):2098–2108. https://doi.org/10.1074/jbc.REV118.002804
Price-Haywood EG, Burton J, Fort D, Seoane L (2020) Hospitalization and mortality among Black patients and White patients with COVID-19. N Engl J Med 382(26):2534–2543. https://doi.org/10.1056/NEJMsa2011686
Sabirli R, Koseler A, Goren T, Turkcuer I, Kurt O (2021) High GRP78 levels in Covid-19 infection: a case-control study. Life Sci 265:118781. https://doi.org/10.1016/j.lfs.2020.118781
Sun Y, Ge L, Rau MJ, Patton MD, Gallan AJ, Felix JC, Rui H (2020) Sensitive and specific immunohistochemistry protocols for detection of SARS-CoV-2 nucleocapsid and spike proteins in formalin-fixed, paraffin-embedded COVID-19 patient tissues [Protocol]. Protocol Exchange. https://doi.org/10.21203/rs.3.pex-1011/v1
Tang X, Yu Q, Wen X, Qi D, Peng J, He J, ..., Wang D (2020) Circulating exosomes from lipopolysaccharide-induced ARDS mice trigger endoplasmic reticulum stress in lung tissue. Shock 54(1) 110-118. https://doi.org/10.1097/SHK.0000000000001397
Tao S, Chen L, Song J, Zhu N, Song X, Shi, R, ..., Zhang Y (2019) Tanshinone IIA ameliorates diabetic cardiomyopathy by inhibiting Grp78 and CHOP expression in STZ-induced diabetes rats. Exp Ther Med, 18(1), 729-734. https://doi.org/10.3892/etm.2019.7580
Tsai YL, Zhang Y, Tseng CC, Stanciauskas R, Pinaud F, Lee AS (2015) Characterization and mechanism of stress-induced translocation of 78-kilodalton glucose-regulated protein (GRP78) to the cell surface. J Biol Chem 290(13):8049–8064. https://doi.org/10.1074/jbc.M114.618736
Wu J, Wu Y, Lian X (2020) Targeted inhibition of GRP78 by HA15 promotes apoptosis of lung cancer cells accompanied by ER stress and autophagy. Biol Open. https://doi.org/10.1242/bio.053298
Yoshida H, Matsui T, Yamamoto A, Okada T, Mori K (2001) XBP1 mRNA is induced by ATF6 and spliced by IRE1 in response to ER stress to produce a highly active transcription factor. Cell 107(7):881–891. https://doi.org/10.1016/s0092-8674(01)00611-0
Zamorano Cuervo N, Grandvaux N (2020) ACE2: Evidence of role as entry receptor for SARS-CoV-2 and implications in comorbidities. Elife 9. https://doi.org/10.7554/eLife.61390
Zhang JL, Li WX, Li Y, Wong MS, Wang YJ, Zhang Y (2021) Therapeutic options of TCM for organ injuries associated with COVID-19 and the underlying mechanism. Phytomedicine 85:153297. https://doi.org/10.1016/j.phymed.2020.153297
Acknowledgements
The authors wish to acknowledge assistance from Christina Gara and David Zimmerman in coordination of data acquisition and analysis, and Mary Rau and Mollie Patton of the Medical College of Wisconsin Tissue Bank for help with acquisition of COVID-19 and non-COVID-19 control specimens and data.
Funding
The funding for this project to IJB was provided by the Bruce and Janine Smith Family, NIH Director's Pioneer Award (NIH grant number- 8DP1HL17650), and the Dean’s Program Development Funds, by the Department of Pathology, Medical College of Wisconsin to HR, and ERJ was supported by Merit Review BX003833. The Children’s Research Institute and the Department of Pediatrics supported PS.
Author information
Authors and Affiliations
Contributions
AP conducted experiments, analyzed data, and wrote the manuscript. ERJ conceived the study, provided patient materials, analyzed and interpreted data, and wrote the manuscript. YS provided lung pathologist expertise, reviewed images, and read and edited the manuscript. JF performed autopsies of decedents. LG performed experiments related to immunohistochemistry. SL provided materials for immunohistochemistry. RN provided critical comments on the manuscript. PN provided critical comments on the manuscript. PS analyzed, interpreted, and wrote the manuscript. HR conceived the study, conducted experiments, analyzed data, and wrote the manuscript. IJB conceived the study, analyzed data, and wrote the manuscript. All authors read and provided feedback on the study.
Corresponding authors
Ethics declarations
Ethics and approval
The Institutional Review Board for Froedtert Hospital & Medical College of Wisconsin granted approval on August 19, 2020, for a waiver of HIPAA authorization requirements at 45 CFR 164 tor review records for this project. All decedent data was approved for access in accordance with 45 CFR 164.512., Project Title: Modeling Health Outcomes using HSP Biosignatures in COVID-19 High Risk Populations.
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Puzyrenko, A., Jacobs, E.R., Sun, Y. et al. Pneumocytes are distinguished by highly elevated expression of the ER stress biomarker GRP78, a co-receptor for SARS-CoV-2, in COVID-19 autopsies. Cell Stress and Chaperones 26, 859–868 (2021). https://doi.org/10.1007/s12192-021-01230-4
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12192-021-01230-4