Abstract
This work has developed an optimized workflow for the targeted analysis of triacylglycerols (TAGs) in milk by liquid chromatography coupled with a Q-Exactive Orbitrap mass spectrometer. First, the effects of resolution (17,500; 35,000; 70,000; 140,000) and automatic gain control target (AGC, from 2×104, 2×105, 1×106, and 3×106) have been optimized with the goal to minimize the injection time, maximize the number of scans, and minimize the mass error. Then, the flow rate of the liquid chromatography system was also optimized by maximizing the number of theoretical plates. The resulting optimized parameters consisted of a flow rate of 200 μL/min, mass resolution of 35,000, and AGC target of 2×105. Such optimal conditions were applied for targeted TAG analysis of milk fat extracts. Up to 14 target triglycerides in milk fat were identified performing a data-dependent HPLC-HRMS-MS2 experiment (t-SIM-ddMS2). The findings reported here can be helpful for MS-based lipidomic workflows and targeted milk lipid analysis.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Triacylglycerols (TAGs) are the principal fats in milk. They are composed of three fatty acids (FA) esterified with glycerol. TAGs in cow’s milk are synthesized mainly from just 16 fatty acids, whose combination may generate up to 163 = 4096 possible distinct triglycerides. Among these, 3454 molecular species of TAGs were recently detected (Liu et al. 2020). The great variability in the potential composition of triglycerides may constitute a new and unexplored chemical alphabet that may be able to detect and reconstruct the processing history of milk, such as the type of breeding of cows, their diet, the stage of lactation, and the season from which the milk is derived (Huppertz and Kelly 2009). Although it is widely recognized that the TAG profile determines the physicochemical properties of milk fat (Dimick et al. 1996; Narine and Marangoni, 1999; Smiddy et al. 2012; Tzompa-Sosa et al. 2016), only very few studies on lipid composition of milk focused on TAG composition (Liu et al., 2017a, b).
One reason could be that the determination of TAGs in milk is still complicated nowadays (Indelicato et al. 2017). The analysis is generally based on chromatographic systems coupled with flame ionization (Gutiérrez et al. 2009), or mass spectrometry (MS) detectors (Fontecha et al. 2000). However, the analysis is complicated by the long sample preparation required to quantitatively extract TAGs, including transesterification for gas chromatography, the fast deterioration of the column performance (Lísa et al. 2011, Beccaria et al. 2014, Kadivar et al. 2013), and the low recovery of certain TAGs during pre-processing and analysis (Aparicio and Aparicio-Ruı́z 2000). A few attempts have also investigated the possibility to detect TAGs by liquid chromatography (LC) coupled with detectors like ultraviolet (UV), evaporative light scattering (ELS), and refractive index detector (RID). However, such detectors have demonstrated low sensitivity toward TAGs (Carvalho et al. 2012).
Instead, a recent and promising approach for the analyses of TAGs in milk consists of the use of high-resolution mass spectrometry (HRMS) (Moulard et al. 2011), which has become the state-of-the-art technique for lipidomic studies (da Silva et al. 2021). HRMS can identify the fatty acid residues of each TAG and benefits from the recent advances in instrument sensitivity, mass resolution, and scanning speed (Kaufmann 2020). These advances lead to unprecedented high-accuracy mass measurements with exact mass resolving power (Makarov et al. 2006). Several attempts were made in recent years to improve the acquisition coverage of HPLC-HRMS–based metabolomics, such as data-set-dependent acquisition (Broeckling et al. 2018), nano-LC/nano electrospray MS (Danne-Rasche et al. 2018), and Data-driven Optimization of MS (Huffman et al. 2019) to name a few examples. Furthermore, HRMS detectors, like the Q-Exactive Orbitrap, offer numerous target or non-target workflows, which have found increased applications in milk lipidomics (Liu et al. 2017).
The working principle of the Q-Exactive Orbitrap is shown in Fig. 1. In details, analytes are firstly separated by LC, then ionized at the source of the Q Exactive Orbitrap. Ions are forced to pass through the S-lens, which filters non-charged compounds and impurities. Furthermore, specific ions of interest up to a wide range of ions are filtered by the quadrupole and, finally, transferred to the C-trap. Here, ions are collected and sent to the Orbitrap mass analyzer, where the mass-to-charge ratios (m/z) of the entering ions are accurately determined at high mass resolution (Zubarev and Makarov 2013).
Schematic representation of the ion selecting and detecting process of the Q-Exactive Orbitrap (according to Thermo Fisher Scientific documentation). In the quadrupole, the ions are selected either by setting a mass range or by selected ion monitoring of single ions. In the C-trap, the ions are accumulated set by the AGC target. The ions oscillate in an orbital motion in the Orbitrap, which induces an image current that is detected. The resolution determines the scan rate at which ions are detected. Higher resolution can increase mass accuracy and sensitivity but decrease the data acquisition rate in an inversely proportional relation. The HCD collision cell fragments the ions if MS2 experiments are performed
Among the most important parameters that must be optimized, is the automatic gain control (AGC) target. This parameter sets the maximum number of ions that can be accumulated in the C-trap before they are transferred into the Orbitrap mass analyzer (Kalli et al., 2013). A further parameter is the maximum injection time (max IT). This sets the time limit after which the collected ions in the C-trap are transferred into the mass analyzer, even when the AGC target is not reached (Kalli et al., 2013). Finally, the third parameter to optimize is the ability to distinguish two peaks of slightly different mass-to-charge ratios in a mass spectrum (Zubarev and Makarov 2013). Overall, mass resolution, AGC target, and max IT greatly affect the resulting scan rate (i.e., number of spectra acquired per time unit) and resolving power of the instrument (Nie et al. 2016). Accordingly, their optimization is pivotal to achieve the best analytical performance. Only few studies attempted the systematic optimization of LC-MS acquisition parameters for lipidomics (Hutchins et al. 2019).
Accordingly, this study aims to optimize the ion detection of TAGs in milk samples. TAGs were chosen, as they represent the most abundant lipid species in milk. The proposed workflow aims to optimize ion acquisition and peak detection during MS analysis of milk lipids. The analytical standard trimargarin was used as model compound to optimize LC and MS parameters. Finally, the workflow was successfully applied for identifying 14 TAGs in milk fat samples.
Materials and Methods
Chemicals and Reagents
Trimargarin, LC-MS-grade formic acid and ammonium formate were purchased from Sigma Aldrich (Steinheim, Germany). LC-MS-grade methanol and acetonitrile were purchased from Honeywell (Selze, Germany), and LC-MS-grade 2-propanol and methyl tert-butyl ether (MTBE) were purchased from Merck KGaA (Darmstadt, Germany) and not purified further. For HPLC methods, ultrasonicated Milli-Q water was employed.
Standard Preparation
The trimargarin triglyceride TG 51:0 (TG(17:0/17:0/17:0)) was used for method optimization. First, 6.52 mg of standard were accurately weighed, transferred to a 20-mL volumetric flask, and brought to volume with 2-propanol to prepare the stock solution for method validation. The stock solution was diluted with methanol/2-propanol (50/50 v/v) to a final concentration of 0.48 μM to be used as working solution and filtered with a 0.45-μm syringe filter to prevent eventual clogging of the instrumentation. Dilutions in the range from 0.095 to 0.575 μM were prepared from the stock solution for the calibration curve and injected in triplicate.
Samples
The samples consisted of whole raw milk provided by the Milk Federation of South Tyrol (Sennereiverband Südtirol). Fresh and unpasteurized bulk milk samples were collected from mountain dairy farms located in the north of Italy (South Tyrol, Italy). The samples were stored at −80°C until analysis.
Sample Preparation
The milk samples were thawed at 8°C overnight. The samples were pooled, carefully mixed, and six aliquots were taken. Fat extraction from the aliquots was carried out according to Breitkopf et al. (2017) based on the extraction method by Matyash et al. (2008) with modifications. In short, 200 μL of milk was mixed with 1.5 mL methanol and vortexed for 1 min. Then, 5 mL of MTBE was added and shaken at 200 rpm for 1 h at room temperature. Then, 1.2 mL of water was added and vortexed for 1 min. The mixture was centrifuged for 10 min at 1000×g at room temperature. The upper phase was collected and the bottom phase re-extracted with 2 volume parts of MTBE/methanol/water (10:3:2.5, v/v/v). The combined upper phases were dried under nitrogen flow at room temperature (MultiVap 8; LabTech S.r.l., Milano, Italy). The dried extracts were dissolved in 5 mL methanol/2-propanol (50:50, v/v) and diluted 1:100 with the same solvent mix. Prior to injection, the sample was filtered with a 0.45-μm PTFE syringe filter. When needed, samples were also spiked with 0.48 μM of trimargarin to evaluate the MS performance during measurement.
Optimization of the MS Parameters in Flow Injection Mode
The system consisted of an Ultimate 3000 UHPLC instrument with UV–Vis detector coupled with a Q-Exactive hybrid quadrupole Orbitrap high-resolution mass spectrometer (HRMS) with heated electrospray ionization (HESI) ion source (Thermo Fisher Scientific, Waltham, MA, USA). The same Q-Exactive Orbitrap HRMS without UHPLC was used for the optimization of the MS parameters.
For flow injection analysis, the Ultimate 3000 UHPLC instrument was coupled to the Q-Exactive Orbitrap HRMS instrument via a Rheodyne switch valve with 5 μL sample loop and syringe injector. The UHPLC system delivered a continuous flow of 200 μL/min consisting of a mixture of solvent A (acetonitrile/water 60:40 v/v with 0.1% formic acid and 10 mM ammonium formate) and solvent B (2-propanol/acetonitrile 90:10 v/v with 0.1% formic acid and 10 mM ammonium formate) at isocratic conditions with 83% solvent B. The HESI probe was set as follows: sheath gas flow at 40 (arbitrary units), auxiliary gas flow at 10 (arbitrary units), sweep gas flow at 0 (arbitrary units), spray voltage in positive ionization mode at 4.00 kV, capillary temperature at 300°C, S-lens RF level at 50%, and aux gas temperature at 100°C. To determine the ideal MS parameters, the mass resolutions R = 17,500; 35,000; 70,000; and 140,000 and the automatic gain control (AGC) target values of 2×104, 2×105, 1×105, and 3×106, with a fixed maximum injection time (IT) of 300 ms, were tested. Acquisition was performed in selected ion monitoring (SIM) mode.
Milk Lipid Separation Using High-Performance Liquid Chromatography Coupled to High-Resolution Mass Spectrometry (HPLC-HRMS)
The chromatographic separation was performed following a modified method by Breitkopf et al. (2017) and optimized for TAG elution. In brief, the stationary phase consisted of a C18 column (Accucore RP-MS, 100 mm × 2.1 mm i.d., 2.6 μm particle size; Thermo Fisher Scientific, Waltham, MA, USA) with a security guard cartridge system (Thermo Fisher Scientific). The mobile phase consisted of a combination of solvent A (acetonitrile/water, 60:40 v/v with 0.1% formic acid and 10 mM ammonium formate) and solvent B (2-propanol/acetonitrile, 90:10, v/v with 0.1% formic acid and 10 mM ammonium formate). The gradient at a flow rate of 200 μL/min was set as follows: 70% B (v/v) for 2 min, then from 70% B to 83% B at 3 min, hold until 8 min then to 84% B at 13 min and hold until 14 min. Sample injection volume was 5 μL using an autosampler with a 20-μL injection loop. After each sample, a wash step with a blank (2-propanol) was introduced with the same chromatographic set-up as before but with a different gradient: from 84% B at 0 min to 97% B at 2 min, hold 97% until 7 min, from 97% at 7 min to 70% B at 8 min followed by a re-equilibration step (70% B) from 8 to 10 min. Blank injection volume was 20 μL. During the wash and re-equilibration step, the flow from the HPLC was diverted to waste using a Rheodyne switch valve, while a flow of 3 μL/min 2-propanol was delivered to the MS using an infusion syringe pump (Thermo Fisher Scientific) to avoid clogging and minimize carry-over effects.
For the MS parameters, the same HESI probe settings were used as described above. Three different MS experiments were used to acquire the standard and sample spectra using the optimized conditions. Full MS experiments were performed in a scan range from 150 to 1500 m/z with a resolution of 35,000 (at m/z 200), an AGC target of 2×105, and a maximum IT of 200 ms. Targeted SIM (t-SIM) experiments were performed with a resolution of 35,000, AGC target of 2×105, max IT of 125 ms, and an isolation window of 4 m/z. Finally, t-SIM–data dependent (dd) MS2 experiments were performed with a resolution of 35,000, an AGC target of 2×105, and a maximum IT of 125 ms for the MS1 acquisitions, and a resolution of 17,500, an AGC target of 1×105, a maximum IT of 50 ms, a loop count of 5, an isolation window of 4.0 m/z, and a normalized stepped collision energy of 15, 30, and 60 eV, for the MS2 acquisitions. For both the t-SIM and t-SIM-ddMS2 experiments, an inclusion list containing the m/z of the targeted molecules to acquire and fragment was used.
Data Processing and Statistical Analysis
The analytical standard was injected in triplicate for each HPLC-HRMS condition and six times for the sample extracts. Data acquisition was performed with tune and Xcalibur software (Thermo Fisher Scientific). Correlation of chemical compounds relative abundances and integration of the area under each peak (HPLC-HRMS XIC integrations) was done using Xcalibur Quan Browser software (Thermo Fisher Scientific) and the LIPIDMAPS lipidomic data base. The limit of detection and quantification (LOD and LOQ), as well as the precision and sensitivity, the latter being the slope of the regression line, were calculated for the trimargarin standard in solution from the regression line after calibration in agreement with Miller and Miller (2018). The obtained LOD and LOQ were theoretically calculated and not experimentally measured. Intraday repeatability was measured as relative standard deviation (RSD %) for the integrated areas of the major base peaks in the six replicates of the extracts. Mass error was calculated by dividing the difference of the actual mass and the theoretical mass by the theoretical mass and expressing in Δppm. The analysis of variance was conducted using XLSTAT annual version 2021.1.1 1092 (Addinsoft 2021, New York, NY, USA).
Results and Discussion
Optimization of HRMS Parameters by Flow Injection Analysis of Trimargarin
Standard solutions of trimargarin were analyzed by a flow injection system (200 μL/min) coupled with a Q-Exactive Orbitrap analyzer, working in positive selected ion monitoring (SIM) mode. A standard of trimargarin was selected as TAG standard for lipidomics. The ammonium adduct [M+NH4]+ of trimargarin with a theoretical m/z of 866.8171 was determined. The injection of 0.48 μM of trimargarin (sample injection of 5 μL) resulted in a peak height of 2.24 ± 0.34 × 106 counts and peak width of 7.76 ± 0.77 s.
Optimization of the MS parameters was next obtained by testing four different mass resolutions R (17,500; 35,000; 70,000; and 140,000 at m/z 200) and four automatic gain control (AGC) values (2×104, 2×105, 1×106, and 3×106). Accordingly, 16 independent experiments were designed, each tested in triplicate (n = 48). For each experimental condition, three parameters were determined: the injection time (IT) needed to acquire one spectrum, the number of scans per peak, and the mass accuracy (i.e., expressed as mass error, Δppm). In all experiments, a maximal injection time of 300 ms was set. This means that when the time to reach the desired AGC target value exceeds such threshold, a mass spectrum is still generated.
The results were examined by a two-way ANOVA. Both mass resolution and AGC target values show a significant effect on the resulting injection time (Fig. 2a), the number of scans per peak width (Fig. 2b), and the mass error (Fig. 2c) (Tukey HSD, p < 0.0001).
In details, Fig. 2a shows the effect of the mass resolution and AGC target values on the resulting number of acquired scans. Resolution was the most important factor (explaining 70% of the total variance), followed by AGC target (22%) and the combination of the two (7%). The interaction between resolution and AGC target was significant (F(1,2), = 170, p < 0.0001). At low resolutions (i.e., 17,500), the selection of the AGC target values was very important for assuring the highest number of scans, which was achieved at the lowest AGC values. Instead, at higher resolutions, the choice of the AGC target value was less important and negligible at 140,000. Similar conclusions were observed in proteomics (Michalski et al. 2011). This result is expected since the AGC target value controls the ion population which is accumulated in the C-trap before being injected into the Orbitrap mass analyzer. So, in general, a higher AGC target value is desired because it increases the ion population. However, at very high resolutions, the number of scans becomes small regardless of the AGC target value chosen.
Similarly, Fig. 2b shows the effect of the mass resolution and AGC target values on the injection time. Again, the main factors that affect the injection time are the mass resolution (explaining 57% of the total variance), followed by the AGC target value (43%) and the combination of the two (9%). The interaction between resolution and AGC target was significant (F(1,2) = 2081, p < 0.0001). At lower resolutions (i.e., 17,500), lower AGC target values led to fast injection times. Kalli et al. (2011) also observed higher injection times with the increase of AGC target for protein detection with the LQT-Orbitrap. Trap filling time was also described by da Silva et al. (2021) to be critical for sensitivity and saturation of lipidomic coverage. Instead, at higher resolutions, the effect of the AGC target was less significant. Ultimately, the AGC target value had no effect on the injection time at a resolution of 140,000. This result is expected as higher resolutions generally requires longer times of acquisition, which limit the number of data points that can be collected across a peak.
Finally, Fig. 2c shows the effect of the mass resolution and AGC target values on the mass accuracy. Here, the main factor was the mass resolution (explaining 75% of the total variance), followed by the combination of AGC target and resolution (14%) and the AGC target (10%). As for the other variables, the interaction between resolution and AGC target was significant (F(1,2) = 12, p < 0.0001). As expected, the highest mass error was observed at the lowest resolution (i.e., 17,500). Conversely, increasing the resolutions, the mass accuracy greatly improved. However, it should be noted that the best mass accuracy could be achieved only with the lowest AGC target value. This result can be explained considering that high AGC target values promote longer injection times and the collection of large ion populations. This may lead to charge effects, which affect the ion distribution inside the Orbitrap and, ultimately, the mass accuracy (Makarov et al. 2006).
Optimal mass analyzer parameters were obtained by selecting those conditions that maximized the number of scans per peak width, minimized the mass error, and guaranteed best exploitation of the injection time (Huang et al., 2021). A desirability function was developed (Candioti et al. 2014) and the best conditions were a resolution R of 35,000, with an AGC target value of 2×105. These conditions led to the highest possible number of scans per peak width (4.58 scans/s) with the smallest mass error (0.16 ppm) and in the shortest injection time. The second-best performance was achieved with a resolution R = 70,000 and with an AGC target value of 2×104, which led to 2.71 scans/s (number of scans per peak width) and a mass error of 0.162 ppm. Instead, the resolution of 140,000 led to the lowest number of scans, likely because of longer scan cycle times (Kalli and Hess 2012). Fewer data points across chromatographic peaks were also observed by Carlsson et al. (2022) when working with polarity switching which increases the cycle time of HRMS measurements. This underlines the importance to adjust scan cycle times depending on the resolution and application needed. In the case of targeted TAG acquisition, at 140,000 resolution, the time for collecting ions exceeded the maximum injection time allowed. This, in practice, resulted in insufficient scans per peak width for all the AGC target values. Conversely, the resolution at 17,500 led to a sufficient saturation of the scan cycle but with insufficient mass resolution.
Accordingly, all the next experiments were based on two optimal sets of experimental conditions, respectively, with R = 35,000 with AGC target = 2×105 and R = 70,000 with AGC target = 2×104.
In-Column HPLC Flow Rate Optimization
The next step was the optimization of the chromatographic separation. The use of a column may limit the speed and depth of MS data acquisition (Huang et al. 2021), and may be critical to the MS efficiency (Shishkova et al. 2016). Therefore, two MS conditions were used, respectively, A: mass resolution R = 70,000 with AGC target value of 2×104, and B: mass resolution R = 35,000 with an AGC target value of 2×105. These two optimized conditions were used to study the effect of flow rate (150, 200, 250, and 300 μL/min) on the resulting peak height, peak width, number of theoretical plates, and number of scans for the analysis of trimargarin.
Based on the results in Table 1, the flow rate greatly affected the retention time, the peak width, the number of theoretical plates, and the number of scans (p < 0.0001). Low flow rates led to lower peak heights, but very high number of scans (p < 0.0001). This can be easily explained considering that a lower flow rate allows a prolonged scan cycle time (Kalli and Hess 2012).
Similar conclusions could be derived at higher resolution (i.e., 70,000), although it should be noted that, when the flow rate is too high (300 μL/min) or too low (i.e., 150 μL/min), the number of ions collected is not sufficient to achieve the best number of scans. Instead, optimal performance was achieved at 200 μL/min, with a significant difference from the other flow rates (Tukey HSD, p < 0.0001).
HPLC-HRMS Method Validation
Trimargarin standards were injected in the HPLC system at a concentration range from 0.191 to 0.575 μM. For each concentration, an extracted ion chromatogram (XIC) was generated (Fig. 3). The peak area versus concentration was plotted in Fig. 3 (inset). With a resolution of 35,000, the calibration curve gave a R2 of 0.996 ± 0.006, limit of detection (LOD) of 0.06 ± 0.02 μM, sensitivity of 6.67 ± 0.31 × 107, and precision of 2% RSD (Table 2). Instead, with a resolution of 70,000, the quantitative analytical performance worsened, showing higher LOD (0.08 ± 0.03 μΜ), lower sensitivity (5.22 ± 0.17 × 107), and lower precision (4% RSD). This result can be explained considering that higher mass resolutions led to slower scan rates and the collection of less data points (Huang et al. 2021). Accordingly, the resolution of 35,000 was chosen for further experiments.
Application of the Optimized Method for Analysis of Milk Fat Extracts
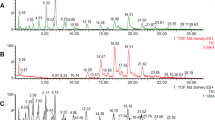
Next, the HPLC-HRMS method with optimized acquisition was applied for the analysis of milk fat extracts. Six aliquots of milk were extracted and analyzed by HPLC-HRMS. Preliminarily, a first MS acquisition was performed in full scan (full-MS) in the range from 150 to 1,500 m/z. This first mode was useful to determine the milk fat profile and choose the ions for SIM-mode acquisition. Figure 4 shows the resulting total ion current (TIC) chromatogram in positive ion mode. Fourteen cluster peaks were observed for the milk lipid profile with the m/z of the base peak of each cluster. The m/z of the most intense ion (base peak) was next used into an inclusion list for targeted SIM (t-SIM) acquisition.
Thereafter, a second acquisition was performed in SIM mode and Fig. 5 shows the resulting XICs. For each peak, 25–60 scans were recorded. For each peak, intraday repeatability of the area under the peak of six replicates of milk fat was always below 5%.
While characterization of isomer TAG species can be achieved with minimal chromatographic separation, if any (Xu et al. 2018; Wang et al. 2016), accurate detection of target lipids from milk can greatly benefit from improved acquisition parameters. Nevertheless, optimization of acquisition parameters for lipidomic studies has found only limited application, which concern especially untargeted analyses (da Silva et al. 2021). Most lipidomics studies on milk are reporting the profiling of the global milk lipidome or that of single lipid classes (Damário et al. 2015; Donato et al. 2011; Li et al. 2017; Liu et al. 2015; Ma et al. 2017; Tzompa-Sosa et al. 2018). Compared to these studies, this work describes the development of a method for optimization of MS parameters aimed to improve the detection and identification of single target lipids in milk.
Identification of Compounds via t-SIM-ddMS2
Identification of the 14 groups of target triglycerides in the milk fat extract was carried out performing a data-dependent HPLC-HRMS-MS2 experiment (t-SIM-ddMS2). The resulting fragmentation spectra were used to characterize the target molecules. For each molecular ion, a chemical formula of the neutral mass was predicted (Table 3). TAG species were identified using the LIPIDMAPS lipidomics database, which classified TAGs based on the total number of carbons of the fatty acid residues (CN, TG x:– ) and the total number of double bonds in the fatty acid residues (DB, TG –:y). For each group of classified TAG molecular species, several fatty acid residues were identified. The combination of those FA resulted in several possible TAGs within one cluster. The final tentative identification was obtained by comparing the resulting fragmentation spectra of each peak with the theoretical spectra generated in LIPIDMAPS (Table 3).
MS response of TAG molecules is knowingly very heterogeneous across species (Holcapek et al., 2005; Han and Gross, 2001). Choosing the correct MS parameters is therefore pivotal for efficient target acquisition. Most studies characterize the milk lipidome without considering the MS performance, which is not much explored. Accordingly, most studies perform lipid profiling rather than single lipid determination (Foroutan et al. 2019, Gresti et al. 1993, Liu et al., 2017a, b, Tzompa-Sosa et al., 2016, Sokol et al. 2015). One of the possible reasons for this is a limited amount of TAG standards available on the market. There are not enough TAG standards available to cover all TAG groups, making the MS optimization laborious. Our study is showing how the use of even one analytical standard (trimargarin) allows to test the influence of MS parameters on the resulting acquisition of TAGs and develop a method to analyze target TAGs in real milk samples.
Conclusions
In this study, targeted milk TAG analysis was performed with an optimized HPLC-HRMS method. Flow rate, mass resolution, and AGC target were optimized on a Q-Exactive Orbitrap HRMS to exploit its full functionalities. Although the current study is not proposing a thorough characterization of all TAG species present in bovine milk, it provided a solution to optimize the ion acquisition and target identification of TAG groups. Accordingly, a flow rate of 200 μL/min, mass resolution of 35,000, and an AGC target value of 2×105 yielded the best data point acquisition, peak generation, and peak area reproducibility for a trimargarin standard and was applied to a real milk fat extract. With the optimized method, a robust analytical performance was achieved. The number of scans for each peak could be maximized for best collection of data points and good peak area reproducibility. The here presented workflow constitutes a valuable addition for lipidomic research. For future studies, this work provides a tool for the optimized acquisition of biomarkers from milk lipids using HRMS and can find application for further food matrices and research fields.
Change history
22 July 2022
Springer Nature’s version of this paper was updated: Missing Open Access funding information has been added in the Funding Note.
References
Aparicio R, Aparicio-Ruíz R (2000) Authentication of vegetable oils by chromatographic techniques. J Chromatogr A 881:93–104. https://doi.org/10.1016/S0021-9673(00)00355-1
Beccaria M, Sullini G, Cacciola F, Donato P, Dugo P, Mondello L (2014) High performance characterization of triacylglycerols in milk and milk-related samples by liquid chromatography and mass spectrometry. J Chromatogr A 1360:172–187. https://doi.org/10.1016/j.chroma.2014.07.073
Breitkopf B, Ricoult SJH, Yuan M, Xu Y, Peake DA, Manning BD, Asara JM (2017) A relative quantitative positive/negative ion switching method for untargeted lipidomics via high resolution LC-MS/MS from any biological source. Metabolomics 13(3):1–21. https://doi.org/10.1007/s11306-016-1157-8
Broeckling SD, Hoyes E, Richardson K, Brown JM, Prenni JE (2018) Comprehensive tandem-mass-spectrometry coverage of complex samples enabled by data-set-dependent acquisition. Anal Chem 90(13):8020–8027. https://doi.org/10.1021/acs.analchem.8b00929
Candioti LV, de Zan MM, Cámara MS, Goicoechea HC (2014) Experimental design and multiple response optimization. Using the desirability function in analytical methods development. Talanta 124:123–138. https://doi.org/10.1016/j.talanta.2014.01.034
Carlsson H, Vaivade A, Khoonsari PE, Burman J, Kultima K (2022) Evaluation of polarity switching for untargeted lipidomics using liquid chromatography coupled to high resolution mass spectrometry. J Chromatogr B 1195:123200. https://doi.org/10.1016/j.jchromb.2022.123200
Carvalho MS, Mendonça MA, Pinho DMM, Resck IS, Suarez PAZ (2012) Chromatographic analyses of fatty acid methyl esters by HPLC-UV and GC-FID. J Braz Chem Soc 23:763–769. https://doi.org/10.1590/S0103-50532012000400023
da Silva KM, Iturrospe E, Heyrman J, Koelmel JP, Cuykx M, Vanhaecke T, Covaci A, van Nuijs ALN (2021) Optimization of a liquid chromatography-ion mobility-high resolution mass spectrometry platform for untargeted lipidomics and application to HepaRG cell extracts. Talanta 235:122808. https://doi.org/10.1016/j.talanta.2021.122808
Damário N, Oliveira DN de, Ferreira MS, Delafiori J, Catharino RR (2015) Cheese lipid profile using direct imprinting in glass surface mass spectrometry. Anal Methods 7:2877–2880. https://doi.org/10.1039/C4AY01899K
Danne-Rasche N, Coman C, Ahrends R (2018) Nano-LC/NSI MS refines lipidomics by enhancing lipid coverage, measurement sensitivity, and linear dynamic range. Anal Chem 90(13):8093–8101. https://doi.org/10.1021/acs.analchem.8b01275
Dimick PS, Reddy SY, Ziegler GR (1996) Chemical and thermal characteristics of milk-fat fractions isolated by a melt crystallization. J Am Oil Chem Soc 73:1647–1652. https://doi.org/10.1007/BF02517966
Donato P, Cacciola F, Cichello F, Russo M, Dugo P, Mondello L (2011) Determination of phospholipids in milk samples by means of hydrophilic interaction liquid chromatography coupled to evaporative light scattering and mass spectrometry detection. J Chromatogr A 1218:6476–6482. https://doi.org/10.1016/j.chroma.2011.07.036
Fontecha J, Ríos J, Lozada L, Fraga M, Juárez M (2000) Composition of goat's milk fat triglycerides analysed by silver ion adsorption-TLC and GC–MS. Int Dairy J 10:119–128. https://doi.org/10.1016/S0958-6946(00)00026-1
Foroutan A, Guo AC, Vazquez-Fresno R, Lipfert M, Zhang L, Zheng J, Badran H, Budinski Z, Mandal R, Ametaj BN, Wishart DS (2019) Chemical composition of commercial cow's milk. J Agric Food Chem 67:4897–4914. https://doi.org/10.1021/acs.jafc.9b00204
Gresti J, Bugaut M, Maniongui C, Bezard J (1993) Composition of molecular species of triacylglycerols in bovine milk fat. J Dairy Science 76:1850–1869. https://doi.org/10.3168/jds.S0022-0302(93)77518-9
Gutiérrez R, Vega S, Díaz G, Sánchez J, Coronado M, Ramírez A, Pérez J, González M, Schettino B (2009) Detection of non-milk fat in milk fat by gas chromatography and linear discriminant analysis. J Dairy Science 92:1846–1855. https://doi.org/10.3168/jds.2008-1624
Han X, Gross RW (2001) Quantitative analysis and molecular species fingerprinting of triacylglyceride molecular species directly from lipid extracts of biological samples by electrospray ionization tandem mass spectrometry. Anal Biochem 295:88–100. https://doi.org/10.1006/abio.2001.5178
Holcapek M, Lísa M, Jandera P, Kabátová N (2005) Quantitation of triacylglycerols in plant oils using HPLC with APCI-MS, evaporative light-scattering, and UV detection. J Separation Science 28:1315–1333. https://doi.org/10.1002/jssc.200500088
Huang P, Liu C, Gao W, Chu B, Cai Z, Tian R (2021) Synergistic optimization of liquid chromatography and mass spectrometry parameters on Orbitrap Tribrid mass spectrometer for high efficient data-dependent proteomics. J Mass Spec 56:e4653. https://doi.org/10.1002/jms.4653
Huffman RG, Chen A, Specht H, Slavov N (2019) DO-MS: data-driven optimization of mass spectrometry methods. J Prot Res 18(6):2493–2500. https://doi.org/10.1021/acs.jproteome.9b00039
Huppertz T, Kelly AL (2009) Properties and constituents of cow's milk. In: Tamime AY (ed) Milk processing and quality management. Wiley-Blackwell Pub./Society of Dairy Technology, Chichester, U.K, Malden, MA, pp 23–47
Hutchins PD, Russell JD, Coon JJ (2019) Accelerating lipidomic method development through in silico simulation. Anal Chem 91(15):9698–9706. https://doi.org/10.1021/acs.analchem.9b01234
Indelicato S, Bongiorno D, Pitonzo R, Di Stefano V, Calabrese V, Indelicato S, Avellone G (2017) Triacylglycerols in edible oils: determination, characterization, quantitation, chemometric approach and evaluation of adulterations. J Chromatogr A 1515:1–16. https://doi.org/10.1016/j.chroma.2017.08.002
Kadivar S, de Clercq N, Nusantoro BP, Le TT, Dewettinck K (2013) Development of an offline bidimensional high-performance liquid chromatography method for analysis of stereospecific triacylglycerols in cocoa butter equivalents. J Agric Food Chem 61:7896–7903. https://doi.org/10.1021/jf401566c
Kalli A, Hess S (2012) Effect of mass spectrometric parameters on peptide and protein identification rates for shotgun proteomic experiments on an LTQ-orbitrap mass analyzer. Proteomics 12:21–31. https://doi.org/10.1002/pmic.201100464
Kalli A, Smith GT, Sweredoski MJ, Hess S (2013) Evaluation and optimization of mass spectrometric settings during data-dependent acquisition mode: focus on LTQ-Orbitrap mass analyzers. J Proteome Res 12:3071–3086. https://doi.org/10.1021/pr3011588
Kaufmann A (2020) High-resolution mass spectrometry for bioanalytical applications: is this the new gold standard? J Mass Spec 55:e4533. https://doi.org/10.1002/jms.4533
Li Q, Zhao Y, Zhu D, Pang X, Liu Y, Frew R, Chen G (2017) Lipidomics profiling of goat milk, soymilk and bovine milk by UPLC-Q-exactive orbitrap mass spectrometry. Food Chem 224:302–309. https://doi.org/10.1016/j.foodchem.2016.12.083
Lísa M, Netušilová K, Franěk L, Dvořáková H, Vrkoslav V, Holčapek M (2011) Characterization of fatty acid and triacylglycerol composition in animal fats using silver-ion and non-aqueous reversed-phase high-performance liquid chromatography/mass spectrometry and gas chromatography/flame ionization detection. J Chromatogr A 1218:7499–7510. https://doi.org/10.1016/j.chroma.2011.07.032
Liu Z, Moate P, Cocks B, Rochfort S (2015) Comprehensive polar lipid identification and quantification in milk by liquid chromatography–mass spectrometry. J Chromatogr B 978-979:95–102. https://doi.org/10.1016/j.jchromb.2014.11.036
Liu Z, Ezernieks V, Wang J, Arachchillage NW, Garner JB, Wales WJ, Cocks BG, Rochfort S (2017a) Heat stress in dairy cattle alters lipid composition of milk. Sci Rep 7:961. https://doi.org/10.1038/s41598-017-01120-9
Liu Z, Wang J, Cocks BG, Rochfort S (2017b) Seasonal variation of triacylglycerol profile of bovine milk. Metabolites. 7(2):24. https://doi.org/10.3390/metabo7020024
Liu Z, Li C, Pryce J, Rochfort S (2020) Comprehensive characterization of bovine milk lipids: triglycerides. ACS omega 5:12573–12582. https://doi.org/10.1021/acsomega.0c01841
Ma L, MacGibbon AK, Mohamed J, Hamid JB, Loy S, Rowan A, McJarrow P, Fong BY (2017) Determination of phospholipid concentrations in breast milk and serum using a high performance liquid chromatography–mass spectrometry–multiple reaction monitoring method. Int Dairy J 71:50–59. https://doi.org/10.1016/j.idairyj.2017.03.005
Makarov A, Denisov E, Kholomeev A, Balschun W, Lange O, Strupat K, Horning S (2006) Performance evaluation of a hybrid linear ion trap/orbitrap mass spectrometer. Anal Chem 78:2113–2120. https://doi.org/10.1021/ac0518811
Matyash V, Liebisch G, Kurzchalia TV, Shevchenko A, Schwudke D (2008) Lipid extraction by methyl-tert-butyl ether for high-throughput lipidomics. J Lipid Res. 49(5):1137–1146. https://doi.org/10.1194/jlr.D700041-JLR200
Michalski A, Cox J, Mann M (2011) More than 100,000 detectable peptide species elute in single shotgun proteomics runs but the majority is inaccessible to data-dependent LC-MS/MS. J Proteome Res 10:1785–1793. https://doi.org/10.1021/pr101060v
Miller J, Miller J (2018) Statistics and chemometrics for analytical chemistry, 6th edn. Pearson education, Harlow, pp 125–134
Moulard Y, Bailly-Chouriberry L, Boyer S, Garcia P, Popot M-A, Bonnaire Y (2011) Use of benchtop exactive high resolution and high mass accuracy orbitrap mass spectrometer for screening in horse doping control. Analytica Chimica Acta 700:126–136. https://doi.org/10.1016/j.aca.2011.01.006
Narine SS, Marangoni AG (1999) Relating structure of fat crystal networks to mechanical properties. Food Res Int 32:227–248. https://doi.org/10.1016/S0963-9969(99)00078-2
Nie L, Zhu M, Sun S, Zhai L, Wu Z, Qian L, Tan M (2016) An optimization of the LC-MS/MS workflow for deep proteome profiling on an Orbitrap Fusion. Anal Methods 8:425–434. https://doi.org/10.1039/C5AY01900A
Shishkova E, Hebert AS, Coon JJ (2016) Now, more than ever, proteomics needs better chromatography. Cell Systems 3:321–324. https://doi.org/10.1016/j.cels.2016.10.007
Smiddy MA, Huppertz T, van Ruth SM (2012) Triacylglycerol and melting profiles of milk fat from several species. International Dairy Journal 24:64–69. https://doi.org/10.1016/j.idairyj.2011.07.001
Sokol E, Ulven T, Færgeman NJ, Ejsing CS (2015) Comprehensive and quantitative profiling of lipid species in human milk, cow milk and a phospholipid-enriched milk formula by GC and MS/MSALL. European Journal of Lipid Science and Technology 117:751–759. https://doi.org/10.1002/ejlt.201400575
Tzompa-Sosa DA, Ramel PR, van Valenberg HJF, van Aken GA (2016) Formation of β polymorphs in milk fats with large differences in triacylglycerol profiles. J Agric Food Chem 64:4152–4157. https://doi.org/10.1021/acs.jafc.5b0573
Tzompa-Sosa DA, Meurs PP, van Valenberg HJF (2018) Triacylglycerol profile of summer and winter bovine milk fat and the feasibility of triacylglycerol fragmentation. Eur. J. Lipid Sci. Technol. 120:1700291. https://doi.org/10.1002/ejlt.201700291
Wang M, Wang C, Han RH, Han X (2016) Novel advances in shotgun lipidomics for biology and medicine. Prog Lipid Res. 61:83–108. https://doi.org/10.1016/j.plipres.2015.12.002
Xu L, Wang X, Jiao Y, Liu X (2018) Assessment of potential false positives via orbitrap-based untargeted lipidomics from rat tissues. Talanta 178:287–293. https://doi.org/10.1016/j.talanta.2017.09.046
Zubarev RA, Makarov A (2013) Orbitrap mass spectrometry. Anal Chem 85:5288–5296. https://doi.org/10.1021/ac4001223
Acknowledgments
This research is funded by the FESR-EFRE project Heumilch (FESR1129 CUP: H36H19000000007, EFRE 2014–2020 “Investitionen in Wachstum und Beschäftigung”). We would like to thank BRING—Beratungsring Berglandwirtschaft, Laimburg Research Center, and the Sennereiverband Südtirol for collaboration and providing the milk samples.
Data Availability Statement
Data available on request from the authors.
Funding
Open access funding provided by Libera Università di Bolzano within the CRUI-CARE Agreement.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Ethical Approval
This article does not contain any studies with human or animal subjects.
Conflict of Interest
The authors declare no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Imperiale, S., Morozova, K., Ferrentino, G. et al. Optimized Identification of Triacylglycerols in Milk by HPLC-HRMS. Food Anal. Methods 15, 2084–2094 (2022). https://doi.org/10.1007/s12161-022-02270-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12161-022-02270-x