Abstract
In the food supply chain, quality control has a very important role in maintaining customer confidence. In the EU, food safety aspects are strictly regulated; however, composition requirements and standard control methods are generally undefined. The rapidly increasing wild boar population has a growing market share in venison or game meat production. Several methods have been described for species identification and control of composition in food products, but only some of these are suitable for routine measurements. The aim of our research was to design a rapid, reliable and simple PCR insertion/deletion (InDel)-based genetic tool suitable for species identification in food quality control laboratories. In total, 59 different swine (Sus scrofa) whole genomes were tested with bioinformatic tools to identify wild boar-specific insertions or deletions. Three independent InDels were suitable for marker development, multiplex PCR amplification and separation in agarose gel. Altogether, 209 samples of wild boar and ten other domestic pig breeds were taken for DNA extraction and validation of the three multiplexed InDel markers. Statistical analysis showed a very high combined predictive value (0.996), indicating the capability of the newly developed markers to detect wild boars with a probability over 99%. Breed assignment tests confirm that the InDel markers developed are suitable for rapid, sensitive and reliable identification of the wild boar meat content of food products. The use of the reported method in food quality control can mean a simple and cost-effective way to maintain consumer confidence and to support the competitiveness of fair producers.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Venison products are highly expensive and premium quality products, therefore, become the target of adulteration. In most European countries, food safety aspects are strictly regulated based on EU legislation; however, composition requirements and standard control methods are generally undefined. Maintaining customer confidence has a key role in the future of venison products, as customers are often wary of quality issues in the case of wild game-derived products. Meat and venison production is also susceptible to food adulteration; thus, the control of identity and origin is the most important factor in quality assurance and traceability. Incorrect labelling represents a commercial issue, as forgers may get a competitive advantage over fair producers (Fajardo et al. 2007; Ballin and Lametsch 2008), and incorrectly identified products can cause health problems for consumers living with sensitivity to various allergens (Pascal and Mahé 2001).
Wild boar (Sus scrofa) is one of the most widely distributed mammals in the world. A significant and continuous increase in their population numbers has been reported by several studies in recent decades (Massei and Genov 2004; Bieber and Ruf 2005; Csányi 2014; Massei et al. 2015). Due to the efforts to control population size, the number of harvested wild boars tends to increase throughout Europe (Massei et al. 2015) and, as a consequence, the total amount and the market share of wild boar venison are expanding within the food industry.
The application of DNA-based methods for food authentication has gained much attention because of food safety and quality concerns. DNA-based methods have effectively boosted the traceability of many processed food products (Fajardo et al. 2007; Wilkinson et al. 2012), as DNA is more stable under conditions typically associated with food preparation procedures (Arslan et al. 2006).
Some types of DNA markers, such as simple tandem repeats (STR) and single-nucleotide polymorphisms (SNP) have been used in pig for breed assignment purposes (Wilkinson et al. 2012; Zsolnai et al. 2013; Lin et al. 2014). Some of these are also capable of detecting wild boar (Caratti et al. 2010; Wilkinson et al. 2012; Lin et al. 2014). However, both STR- and SNP-based methods also have their disadvantages and limitations. The complex and heterogenous mutation pattern of STRs (Ellegren 2004) introduces ambiguities, and stutter bands or technical artefacts (allelic dropouts, null alleles, size homoplasy) can cause genotyping errors (Pompanon et al. 2005). Using SNPs, a large number of loci are needed to get feasible information (Syvänen 2001; Wilkinson et al. 2012); moreover, both genotyping methods are relatively expensive at small or medium scale and require special equipment (Wilkinson et al. 2012; Lin et al. 2014).
One of the most popular recent methods is real-time PCR amplification in food composition studies due to its accuracy, reliability and the potential of multiplex measurements (Fajardo et al. 2008). Methods routinely used in research environments may have high costs and low throughput. The necessity of expensive instrumental and human resources makes most of them unsuitable for routine measurements in quality control laboratories (Lockley and Bardsley 2000).
Short, biallelic molecular markers represent a good alternative for simple species identification assay designs. Insertion/deletion (InDel) markers have the potential of developing high-throughput, cost-effective rapid tests for routine monitoring of meat samples. InDel markers are based on insertion/deletion polymorphisms of certain regions of the genome sequence and can be easily analysed as length differences of PCR products by simple agarose gel electrophoresis. InDel markers have recently been used for selection in animal breeding (Ren et al. 2017; Crespo-Piazuelo et al. 2019), genetic mapping (Väli et al. 2008), diagnostics of genes encoding human diseases like Alzheimer’s disease (Bhattramakki et al. 2002; Lehmann et al. 2005) and identification of humans for forensic purposes (Pereira et al. 2009; Fondevila et al. 2012). Longer insertions/deletions have the potential of species identification, and favourable characteristics make them ideal for the examination of degraded DNA, including analysis in short amplicon size ranges, high multiplexing capability and low mutation rates (Väli et al. 2008; Fondevila et al. 2012; Crespo-Piazuelo et al. 2019). In addition, as length-based markers, InDels can be analysed with the use of fluorescent dye-labelled PCR primers subsequently separated and detected by capillary electrophoresis as standard forensic STR markers.
The development of next-generation sequencing (NGS) technologies has changed many fields of life sciences and improved the detection of genomic variants for genetic marker development. Insertions and deletions have been characterised to a lesser extent than SNPs or STRs, although whole-genome sequence data provide the possibility to find these specific regions (Zhang et al. 2017; Crespo-Piazuelo et al. 2019). For the purpose of this study, the genomes of three Mangalica breeds were sequenced and analysed previously using the abovementioned method (Molnár et al. 2014).
In this study, the main objectives were (1) to identify wild boar-specific insertions/deletions with analytical potentials, (2) to develop and validate markers in these regions and (3) to design a rapid but reliable simple multiplex method suitable for use in food quality control. Our effort to fortify the control of origin may support the competitiveness of fair producers by the maintenance of traceability. Moreover, it also helps to prevent intended crossbreeding that aims to increase productivity with deleterious effects on the genetic structure of wild boar populations.
Materials and Methods
Sampling and DNA Extraction
A total of 209 different muscle, blood and hair samples were collected for analysis (Table 1). Domestic pig samples, collected on farms and abattoirs, represented ten different pig breeds. Mangalica samples from the Hungarian Pig Tissue collection of the research consortium of MANGFOOD were used (Zsolnai et al. 2013). The dataset includes samples of 65 wild boar individuals; muscle samples were collected at various hunting events and venison processing plants, both in different regions of Hungary. Blood samples were collected from living animals into EDTA-coated sampling tubes by a trained veterinarian according to standard veterinary medical practice. Muscle tissue samples were collected from slaughtered animals in sampling tubes containing 96% ethanol. Hair samples were collected from living animals in sampling bags. All samples were stored at − 20 °C until processed in the laboratory.
Total genomic DNA from blood samples was extracted using the Duplicα Prep Automated DNA/RNA Extraction System (EuroClone, Italy); genomic DNA was obtained from muscle tissue using the Genomic DNA Mini Kit (Geneaid, USA) and from hair samples using the QIAamp DNA Investigator Kit (QIAGEN, Germany) according to the manufacturer’s instructions. DNA samples were checked and quantified in a NanoDrop 1000 spectrophotometer (Thermo Fisher Scientific, USA) and stored at − 20 °C until processing.
Identification of InDels
A total of 59 different suid whole genomes (Groenen et al. 2012; Molnár et al. 2014) were downloaded from the public SRA database of NCBI and were compared to identify sequence differences between wild boars and individuals of other breeds. The whole-genome sequencing (WGS) data of 13 wild boars, 4 Mangalicas and 42 other breeds (dominantly Duroc, landrace, large white, Meishan, Pietrain and some less widespread breeds) were obtained. Paired-end sequence reads were mapped against the indexed pig reference genome assembly (Sscrofa10.2) using the Burrows-Wheeler Alignment (BWA) tool (Li and Durbin 2009). Unmapped and non-unique reads were filtered out, and the obtained alignment result files (in bam format) were sorted and indexed using SAMtools (Li et al. 2009). Variant calling was performed with SAMtools mpileup. Species-specific sequence differences were extracted from variant callings using BCFtools (Li et al. 2009) and filtered on the basis of specificity and size. Selected loci were visually checked using integrative genomics viewer (IGV) software (Robinson et al. 2011) on available sequences. Planned primer sequences were verified using UCSC in silico PCR (http://genome.ucsc.edu/cgi-bin/hgPcr).
Marker Development and Genotyping
Wild boar species-specific insertions or deletions within the approximate size of 100–1000 bp were targeted, as this range has the potential of easy identification with agarose gel electrophoresis (AGE). Primer3 (Rozen and Skaletsky 2000) was used to design primers for the identified regions. Selected primers were individually tested on wild boar and domestic pig samples and optimised for multiplex PCR to achieve efficient amplification and fidelity. Two sets of primers were designed on the same regions: one unlabelled for the simple AGE method and a fluorescent-labelled set feasible for rapid and high-throughput screening of a bigger number of samples by capillary electrophoresis.
Unlabelled W Marker Set
Multiplex PCR amplifications were performed in a total volume of 15 μL containing 60-ng template DNA, 0.5 μL of each primer and 1 × QIAGEN Multiplex PCR Master Mix (QIAGEN, Germany). Amplification was conducted in a LifeECO thermal cycler (BIOER, China) using the following cycling conditions: initial activation at 95 °C for 15 min, followed by 40 cycles of denaturation at 94 °C for 40 s, annealing at 60 °C for 40 s and extension at 72 °C for 30 s, with a final extension step at 72 °C for 5 min (detailed primer concentrations and cycling conditions are found in supplementary material Table S1). PCR products were separated initially with AGE using 1.2% agarose gel. GeneRuler 1kb DNA Ladder (Thermo Fisher Scientific, USA) was used as a size standard.
Labelled W Marker Set
The unlabelled primers were redesigned to a maximum of 500 bp length for the fragment-analysis method. The amplifications were performed in a total volume of 25 μL containing 45-ng template DNA, the optimum concentration of each primer and 1 × QIAGEN Multiplex PCR Master Mix (QIAGEN, Germany) (detailed primer concentrations and cycling conditions are found in supplementary material Table S1). PCR conditions were the same as used in the case of the unlabelled set.
Fluorescently labelled PCR products were separated on an ABI 3130xl Genetic Analyzer (16 capillaries, 50 cm length) using BigDye Terminator v3.1 Cycle Sequencing Kit and POP-7 polymer for the separation following 100× dilution. LIZ 500 (Applied Biosystems) was used as an internal standard. Allele designation was made with Peak Scanner v1.0 software (Applied Biosystems).
Genotyping of individuals was done based on gel images using GeneRuler 100 bp+ DNA Ladder (Thermo Fisher Scientific, USA) as size standard; individual genotypes were stored and analysed as allele size tables. Thirteen tetrameric STR markers (Lin et al. 2014) were used to make a comparison with the InDel genetic composition results of the collected samples. Multiplex PCR conditions were partially modified for better amplification; genotyping was done as described by Lin et al. (2014) (adapted primer concentrations and cycling conditions are found in supplementary material Table S2).
Breed Assignments and Statistical Analysis
Allele frequencies, heterozygosity values and genetic indices were calculated using the software GenAlEx (Peakall and Smouse 2012) and custom scripts. Parameters to evaluate the efficiency, such as random match probabilities (RMPs) and diagnostic effectiveness and predictive values for each locus and profile (i.e. accumulated values for the whole marker set), were calculated using well-established formulas following Pereira et al. (2009) and Fondevila et al. (2012).
Several different approaches were used to assess the applicability of the markers for breed assignment purposes. Firstly, the probability of an animal belonging to a certain breed was calculated with the partial Bayesian approach of Rannala and Mountain (1997) implemented in GeneClass2 (Piry et al. 2004), with 10,000 simulated multi-locus genotypes and a threshold for the individual exclusion of 0.01. The Bayesian model-based clustering algorithm of the STRUCTURE software (Pritchard et al. 2000) was also applied, using the admixture model and correlated allele frequencies with no prior breed indicated; ten independent runs for each K from one to ten were performed with 750,000 iterations after a 250,000-iteration burn-in period. To determine the number of genetic clusters, we used the method of Evanno et al. (2005) based on the second-order rate of change in log probability between successive K values as implemented by the program Structure Harvester (Earl and vonHoldt 2012). All breed assignment analyses were made with the InDel and STR marker results separately and then with the entire dataset combined.
Results and Discussion
InDel Detection and Marker Testing
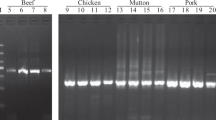
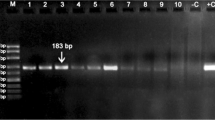
Whole-genome sequencing data of pigs and wild boars was used for InDel detection. The bioinformatic pipeline identified 39,716 loci with specific differences between the genomes of wild boar and other pigs. After initial filtering, five wild boar-specific InDels were selected for further testing in the adequate 100–1000-bp size range. Primer design was successful on all five loci, and the primers designed were tested on different pig and wild boar samples individually. Three of the markers amplified well, with analytical potential, based on AGE images (Fig. 1). These three markers were applicable for easy individual genotyping of samples. Chromosome locations, fragment lengths and primer sequences of the markers used in the multiplex genotyping are shown in Table 2.
The three InDel markers (referred to as W markers hereinafter) were tested on different pig and wild boar samples individually and then combined in a multiplex protocol for efficient daily use application. Different fragment sizes were well separated in gel electrophoresis; thus, different genotypes were easy to identify. Combining the three markers in one reaction gives a rapid result and makes sample identification more cost-effective. AGE images of PCR product sizes and allele lengths for the different genotypes are shown in Fig. 1a and b.
The AGE method alone proved to be feasible for the separation of PCR products, making the genotyping of the DNA samples quite simple. However, fluorescent gel electrophoresis adaptation was implemented for better resolution by labelling the PCR products with fluorescent dyes. Three different fluorescent dyes were used to tag one of the primers of each of the three W markers (Table 2).
Genotyping and Breed Assignment
The pig and wild boar reference set used for characterising the W markers comprised 120 pigs from different breeds and 65 wild boar individuals (Table 1). Samples added for validation (column 3 of Table 1) were not included in the reference genotyping. In order to prove species specificity, genomic samples of other non-suid species occurring in meat products were tested, including cattle (Bos taurus), sheep (Ovis aries), horse (Equus caballus) and two poultry species (Anser anser, Gallus gallus). Interspecies tests gave no PCR products in the expected size range. In silico PCR predicted non-specific products as well when tested on the genomes of turkey (Meleagris gallopavo) and rat (Rattus rattus) (http://genome.ucsc.edu/cgi-bin/hgPcr).
All three InDels amplified efficiently and reliably with detectable products, and all markers proved to be biallelic on the reference samples. The deletion alleles of the selected InDels were not exclusively wild boar specific nor were all the insertions pig specific. But, there were differences between the allele frequencies of wild boars and pigs; thus, the markers are capable of the differentiation of wild boars. The predictive values of markers for detecting wild boars were moderate when tested individually, as it was assumable for biallelic markers. However, the combined predictive value was much higher (0.996), meaning that the use of the three W markers together is capable of detecting wild boars with a probability over 99%, based on our reference sample set. Reference allele frequencies, expected and observed heterozygosity of the individual markers with the individual and combined random match probability, diagnostic effectiveness and predictive values are shown in Table 3. Expected and observed heterozygosity values varied within a wide range (0.117–0.615) and were similar to those described by Lin et al. (2014), but were relatively lower than the findings of Velickovic in the case of a domestic pig as well as wild boar (Table 3).
Bayesian Cluster Analysis in STRUCTURE
Breed assignment was performed using the program STRUCTURE; the Q plot output was used to visualise the clustering results. In the case of W markers, both likelihood scores and the second-order rate of change in log probability indicate the presence of two groups in the sample set (Fig. 2a). This meets our presumption that the W markers are able to distinguish between wild boar and domestic pig samples.
Q plot output of STRUCTURE clustering results: a using the three wild boar-specific markers designed, b using the STR marker set, c using the combined W and STR markers. K, number of clusters; P, Pietrain; H, Hampshire; LW, large white; H39, H39 × large white; L, landrace; D, Duroc; MxD, Mangalica × Duroc; BM, blond Mangalica; SM, swallow-belly Mangalica; RM, red Mangalica; WB, wild boar; WB2, additional wild boar samples used in validation; D2, additional Duroc samples used in validation
Two groups in the population had the highest statistical probability in the case of STR markers as well; however, classification of the different breeds was not consequent; thereby, these markers were not reliable in separating wild boar from other breeds (Fig. 2b). Additionally, we found that in most cases, STRs classified the indigenous Mangalica breeds as wild boar. The underlying reason for this can be that the genetic relationship of Mangalica with wild boar is much closer than that with other breeds, and crossbreeding could occur. Mangalica breeds and the extensive management techniques associated with these pigs (i.e. free herding) allowed crossbreeding with wild boars (Manunza et al. 2016; Frank et al. 2017).
Combining the dataset of W and STR markers had the best reliability to distinguish wild boar from a domestic pig. In this case, the optimal classification had four groups, and the results were highly consequent in identifying wild boars, domestic breeds and crossbreeds/hybrids (Fig. 2c). However, the reliability of W markers alone did not differ significantly from the combined W plus STR results.
Bayesian Cluster Analysis with GeneClass2
With the partial Bayesian approach implemented in GeneClass2, using only the W markers could correctly identify 115 (95.8%) of the 120 domestic pig reference samples as domestic, and 61 (93.9%) of the 65 wild boars as wild boar (assignment score 75<) (Table 4). The remaining four wild boar and five domestic pig samples had assignment scores of equal probability for being domestic or wild (assignment score < 75), and these samples were classified as wild-domestic crossbreeds (Table 4). The clustering results again indicate that the W markers alone were able to distinguish wild boar and domestic pig samples with fairly high reliability (Fig. 3a).
Bayesian clustering results: a using the three wild boar-specific markers designed, b using the STR marker set, c using the combined W and STR markers. K, number of clusters; P, Pietrain; H, Hampshire; LW, large white; H39, H39 × large white; L, landrace; D, Duroc; MxD, Mangalica × Duroc; BM, blond Mangalica; SM, swallow-belly Mangalica; RM, red Mangalica; WB, wild boar; WB2, additional wild boar samples used in validation; D2, additional Duroc samples used in validation
Using the STR marker set for assignment test gave a slightly different result (Fig. 3b). In this case, all wild boar samples were identified as wild, but some of the Mangalica and crossbreeds were also identified as wild boar or as a crossbreed. This can be a consequence of the known genetic connectivity between Mangalica breeds and wild boars, as mentioned before.
Combining the dataset of the W and STR markers gave the best separation, similar to STRUCTURE results, although in this case, all domestic pig samples were identified as domestic and every wild boar as wild (Fig. 3c). However, again as in the case of STRUCTURE, the reliability of W markers alone did not differ significantly from that of the combined W and STR dataset.
In the validation process, a trial test was made with an additional 12 Duroc samples with known pedigree and 12 wild boar samples to test the reliability and consistency of the markers. In the case of the W marker set, both clustering methods classified Duroc samples as domestic and the others as wild boar, as expected (Figs. 2a and 3a). Breed assignment tests confirm the expectation that the designed InDel markers are suitable for the identification of wild boar in meat samples. The identification of hybrids with the multiplex W marker set alone was possible as well. In the case of samples identified as a hybrid, further, higher resolution methods such as SNPs, microsatellites and partial or whole-genome sequencing can provide more information about the genetic background. The STR-based method designed by Lin et al. (2014) has a much higher error, as it could not discriminate between Mangalica and wild boar. The use of microsatellites in the study of Velickovic et al. (2014) gave a better resolution in the investigated populations than that described by Lin et al. (2014), but the use of capillary electrophoresis increases the cost and decreases the throughput compared with the InDel-based method described here using AGE separation. Although the combination of the above described W marker set characterises only 3 shorter but specific regions of the whole genome, the reliability seems much higher than that of methods based on colour genes like MC1R (Fajardo et al. 2008; Fernández et al. 2004) or on mtDNA (Fang and Andersson 2006). Conyers et al. (2012) designed a method based on microsatellite markers, namely 20 simple sequence repeats (SSR) for the differentiation of wild boar from a domestic pig in meat samples, which proved to be feasible for deciding whether or not a meat sample contained wild boar, but unsuitable for the accurate quantification in mixtures; the same can be concluded in case of the W marker set. The reliability of the method based on 20 SSRs was very close to our findings, although it seems to be difficult to use in routine tests as it is quite complex, results cannot be obtained in simple, single-step analysis, and evaluation demands knowledge in the use of various statistical tools (Conyers et al. 2012). In the case of the W marker set sample preparation, PCR amplification and AGE separation can be done easily, and the evaluation does not require complex statistical analysis. This could represent the main benefit of the W marker set designed. Even a higher number of microsatellite markers may have lower potential in discriminating closer related subspecies (as it seemed when using the STR markers on Mangalica and wild boar samples in our study) if the polymorphism of the involved genomic regions is not sufficiently high (Conyers et al. 2012). In our case, the combination of only three specific biallelic InDel markers was sufficient to provide a predictive value higher than 99%, comparable with the methods based on 13 or 20 STRs. SNP chips can be very accurate and specific in identification; their disadvantage is the cost and the complexity of the method and the evaluation, respectively (Ramos et al. 2011). Isothermal ‘on-site’ amplification with test stripe detection of products can be an optional pre-filtering method in the traceability control of products. Szántó-Egész et al. (2016) successfully designed a recombinase polymerase amplification (RPA)-based breed-specific method for the rapid detection of Mangalica meat in food. The W plex InDel marker set would be feasible for the development of a similar rapid field test. This can be a further perspective of our study, as it is not yet adapted for the detection of wild boar in meat. For DNA quantification in meat products, Floren et al. (2015) designed a workflow chart with real-time PCR as a first step followed by a one-step or two-step droplet digital PCR. This ensures that undesired admixtures can be detected even at very low concentrations in a product. Our recommendation would be to include a more simple and fairly rapid InDel-based test as an initial step and to implement quantitative PCR only if this first test is positive.
Conclusions
Identification of wild boar-specific insertions/deletions with bioinformatic tools using whole-genome sequences proved to be successful. We were able to design specific primers for the amplification of three from the five identified InDels. The primers designed could be used successfully in simple multiplex reactions; PCR products were feasible in size for AGE identification. Validation showed that the designed W marker set proved to be a sensitive tool for distinguishing wild boar from other domesticated breeds in meat samples. The autosomal STR marker results had less confidence in separation than the W markers. A combination of the two methods resulted the most reliable method for identification; however, W markers alone also enabled reliable separation. Thus, the newly developed markers have the potential for rapid, sensitive and reliable identification of wild boar meat content of food products in routine laboratory practice. The use of the reported method in food quality control can mean a simple and cost-effective way to maintain consumer confidence and to support the competitiveness of fair producers.
References
Arslan A, Ilhak OI, Calicioglu M (2006) Effect of method of cooking on identification of heat processed beef using polymerase chain reaction (PCR) technique. Meat Sci 72:326–330. https://doi.org/10.1016/j.meatsci.2005.08.001
Ballin NZ, Lametsch R (2008) Analytical methods for authentication of fresh vs. thawed meat–A review. Meat Sci 80(2):151–158. https://doi.org/10.1016/j.meatsci.2007.12.024
Bhattramakki D, Dolan M, Hanafey M, Wineland R, Vaske D, Register JC, Tingey SV, Rafalski A (2002) Insertion-deletion polymorphisms in 3′ regions of maize genes occur frequently and can be used as highly informative genetic markers. Plant Mol Biol 48:539–547. https://doi.org/10.1023/A:1014841612043
Bieber C, Ruf T (2005) Population dynamics in wild boar Sus scrofa: ecology, elasticity of growth rate and implications for the management of pulsed resource consumers. J Appl Ecol 42:1203–1213. https://doi.org/10.1111/j.1365-2664.2005.01094.x
Caratti S, Rossi L, Sona B, Origlia S, Viara S, Martano G, Torre C, Robino C (2010) Analysis of 11 tetrameric STRs in wild boars for forensic purposes. Forensic Sci Int Genet 4:339–342. https://doi.org/10.1016/j.fsigen.2010.07.001
Conyers CM, Allnutt TR, Hird HJ, Kaye J, Chisholm J (2012) Development of a microsatellite-based method for the differentiation of European wild boar (Sus scrofascrofa) from domestic pig breeds (Sus scrofadomestica) in food. J Agric Food Chem 60(13):3341–3347. https://doi.org/10.1021/jf205109b
Crespo-Piazuelo D, Criado-Mesas L, Revilla M, Castelló A, Fernández AI, Folch JM, Ballester M (2019) Indel detection from Whole Genome Sequencing data and association with lipid metabolism in pigs. PLoS ONE 14:e0218862. https://doi.org/10.1371/journal.pone.0218862
Csányi S (2014) Wild boar population dynamics and management in Hungary. J Mountain Ecol 3:222–225 http://www.mountainecology.org/index.php/me/article/view/121
Earl DA, vonHoldt BM (2012) STRUCTURE HARVESTER: a website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv Genet Resour 4:359–361. https://doi.org/10.1007/s12686-011-9548-7
Ellegren H (2004) Microsatellites: simple sequences with complex evolution. Nat Rev Genet 5:435–445. https://doi.org/10.1038/nrg1348
Evanno G, Regnaut S, Goudet J (2005) Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. MolEcol 14:2611–2620. https://doi.org/10.1111/j.1365-294X.2005.02553.x
Fajardo V, González I, López-Calleja I, Martín I, Rojas M, Hernández PE, García T, Martín R (2007) Identification of meats from red deer (Cervuselaphus), fallow deer (Damadama), and roe deer (Capreoluscapreolus) using polymerase chain reaction targeting specific sequences from the mitochondrial 12S rRNA gene. Meat Sci 76:234–240. https://doi.org/10.1016/j.meatsci.2006.11.004
Fajardo V, González I, Martín I, Rojas M, Hernández PE, García T, Martín R (2008) Real-time PCR for detection and quantification of red deer (Cervuselaphus), fallow deer (Damadama), and roe deer (Capreoluscapreolus) in meat mixtures. Meat Sci 79(2):289–298. https://doi.org/10.1016/j.meatsci.2007.09.013
Fang M, Andersson L (2006) Mitochondrial diversity in European and Chinese pigs is consistent with population expansions that occurred prior to domestication. Proc R Soc B Biol Sci 273(1595):1803–1810. https://doi.org/10.1098/rspb.2006.3514
Fernández A, Fabuel E, Alves E, Rodriguez C, Silió L, Óvilo C (2004) DNA tests based on coat colour genes for authentication of the raw material of meat products from Iberian pigs. J Sci Food Agric 84(14):1855–1860. https://doi.org/10.1002/jsfa.1829
Floren C, Wiedemann I, Brenig B, Schütz E, Beck J (2015) Species identification and quantification in meat and meat products using droplet digital PCR (ddPCR). Food Chem 173:1054–1058. https://doi.org/10.1016/j.foodchem.2014.10.138
Fondevila M, Phillips C, Santos C, Pereira R, Gusmao L, Carracedo A, Butler JM, Lareu MV, Vallone PM (2012) Forensic performance of two insertion–deletion marker assays. Int J Legal Med 126:725–737. https://doi.org/10.1007/s00414-012-0721-7
Frank K, Molnár J, Barta E, Marincs F (2017) The full mitochondrial genomes of Mangalica pig breeds and their possible origin. Mitochondrial DNA Part B: Resources 2:730–734. https://doi.org/10.1080/23802359.2017.1390415
Groenen MA, Archibald AL, Uenishi H, Tuggle CK, Takeuchi Y, Rothschild MF, Li S et al (2012) Analyses of pig genomes provide insight into porcine demography and evolution. Nature 491:393–398. https://doi.org/10.1038/nature11622
Lehmann DJ, Cortina-Borja M, Warden DR, Smith AD, Sleegers K, Prince JA, van Duijn CM, Kehoe PG (2005) Large meta-analysis establishes the ACE insertion-deletion polymorphism as a marker of Alzheimer’s disease. Am J Epidemiol 162:305–317. https://doi.org/10.1093/aje/kwi202
Li H, Durbin R (2009) Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 25:1754–1760. https://doi.org/10.1093/bioinformatics/btp324
Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, The 1000 Genome Project Data Processing Subgroup (2009) The sequence alignment/map format and SAMtools. Bioinformatics 25:2078–2079. https://doi.org/10.1093/bioinformatics/btp352
Lin YC, Hsieh HM, Lee JCI, Hsiao CT, Lin DY, Linacre A, Tsai LC (2014) Establishing a DNA identification system for pigs (Sus scrofa) using a multiplex STR amplification. Forensic Sci Int Genet 9:12–19. https://doi.org/10.1016/j.fsigen.2013.10.006
Lockley AK, Bardsley RG (2000) DNA-based methods for food authentication. Trends Food Sci Technol 11(2):67–77. https://doi.org/10.1016/S0924-2244(00)00049-2
Manunza A, Amills M, Noce A, Cabrera B, Zidi A, Eghbalsaied S, Carrillo de Albornoz E, Portell M, Mercadé A, Sànchez A, Balteanu V (2016) Romanian wild boars and Mangalitza pigs have a European ancestry and harbour genetic signatures compatible with past population bottlenecks. Sci Rep 6:29913. https://doi.org/10.1038/srep29913
Massei G, Genov PV (2004) The environmental impact of wild boar. Galemys 16:135–145
Massei G, Kindberg J, Licoppe A, Gačić D, Šprem N, Kamler J, Baubet E, Hohmann U, Monaco A, Ozolins J, Cellina S, Podgórski T, Fonseca C, Makov N, Pokorny B, Rossel C, Náhlik A (2015) Wild boar populations up, numbers of hunters down? A review of trends and implications for Europe. Pest Manag Sci 71:492–500. https://doi.org/10.1002/ps.3965
Molnár J, Nagy T, Stéger V, Tóth G, Marincs F, Barta E (2014) Genome sequencing and analysis of Mangalica, a fatty local pig of Hungary. BMC Genomics 15:761. https://doi.org/10.1186/1471-2164-15-761
Pascal G, Mahé S (2001) Identity, traceability, acceptability and substantial equivalence of food. Cell Mol Biol 47:1329–1342 11838953
Peakall R, Smouse PE (2012) GenAlEx 6.5: genetic analysis in Excel. Population genetic software for teaching and research – an update. Bioinformatics 28:2537–2539. https://doi.org/10.1111/j.1471-8286.2005.01155.x
Pereira R, Phillips C, Alves C, Amorim A, Carracedo Á, Gusmão L (2009) A new multiplex for human identification using insertion/deletion polymorphisms. Electrophoresis 30:3682–3690. https://doi.org/10.1002/elps.200900274
Piry S, Alapetite A, Cornuet JM, Paetkau D, Baudouin L, Estoup A (2004) GENECLASS2: A software for genetic assignment and first-generation migrant detection. J Hered 95:536–539. https://doi.org/10.1093/jhered/esh074
Pompanon F, Bonin A, Bellemain E, Taberlet, P (2005) Genotyping errors: causes, consequences and solutions. Nature Reviews Genetics 6(11): 847–859.
Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155:945–959 10835412
Ramos AM, Megens HJ, Crooijmans RPMA, Schook LB, Groenen MAM (2011) Identification of high utility SNPs for population assignment and traceability purposes in the pig using high-throughput sequencing. Anim Genet 42(6):613–620. https://doi.org/10.1111/j.1365-2052.2011.02198
Rannala B, Mountain JL (1997) Detecting immigration by using multilocus genotypes. Proc Natl Acad Sci U S A 94(17):9197–9201. https://doi.org/10.1073/pnas.94.17.9197
Ren F, Yu S, Chen R, Lv X, Pan CJ (2017) Identification of a novel 12-bp insertion/deletion (indel) of iPS-related Oct4 gene and its association with reproductive traits in male piglets. Anim Reprod Sci 178:55–60. https://doi.org/10.1016/j.anireprosci.2017.01.009
Robinson JT, Thorvaldsdóttir H, Winckler W, Guttman M, Lander ES, Getz G, Mesirov JP (2011) Integrative Genomics Viewer. Nat Biotechnol 29:24–26. https://doi.org/10.1038/nbt.1754
Rozen S, Skaletsky HJ (2000) Primer3 on the WWW for general users and for biologist programmers. In: Krawetz S, Misener S (eds) Bioinformatics Methods and Protocols: Methods in Molecular Biology. Humana Press, Totowa, pp 365–368. https://doi.org/10.1385/1-59259-192-2:365
Syvänen AC (2001) Accessing genetic variation: Genotyping single nucleotide polymorphisms. Nat Rev Genet 2:930–942. https://doi.org/10.1038/35103535
Szántó-Egész R, Jánosi A, Mohr A, Szalai G, Szabó EK, Micsinai A, Sipos R, Rátky J, Anton I, Zsolnai A (2016) Breed-specific detection of Mangalica meat in food products. Food Anal Methods 9(4):889–894. https://doi.org/10.1007/s12161-015-0261-0
Väli Ü, Brandström M, Johansson M, Ellegren H (2008) Insertion-deletion polymorphisms (indels) as genetic markers in natural populations. BMC Genet 9:8. https://doi.org/10.1186/1471-2156-9-8
Velickovic N, Dan M, Ferreira E, KocisTubic N, Obreht D, Beukovic M, Fonseca C (2014) Comparative analysis of wild boar and domestic pig populations based on microsatellites data, Proceedings of the 3th International Symposium on Hunting. Belgrade, Serbia. UDC: 599.731.1:591.526:81’366.55
Wilkinson S, Archibald AL, Haley CS, Megens HJ, Crooijmans RP, Groenen MA, Wiener P, Ogden R (2012) Development of a genetic tool for product regulation in the diverse British pig breed market. BMC Genomics 13:580. https://doi.org/10.1186/1471-2164-13-580
Zhang T, Gu M, Liu Y, Lv Y, Zhou L, Lu H, Liang S, Bao H, Zhao H (2017) Development of novel InDel markers and genetic diversity in Chenopodium quinoa through whole-genome re-sequencing. BMC Genomics 18:685. https://doi.org/10.1186/s12864-017-4093-8
Zsolnai A, Tóth G, Molnár J, Stéger V, Marincs F, Jánosi A, Ujhelyi G, Koppányné Szabó E, Mohr A, Anton I, Szántó-Egész R, Sipos R, Egerszegi I, Dallmann K, Tóth P, Micsinai A, Brüssow KP, Rátky J (2013) Looking for breed differentiating SNP loci and for a SNP set for parentage testing in Mangalica. Archiv Tierzuht 56:200–207. https://doi.org/10.7482/0003-9438-56-019
Acknowledgements
The authors would like to express their gratitude to Villányi-Vad Ltd. and all project participants for providing samples for this study.
Funding
Open access funding provided by National Agricultural Research and Innovation Centre. This study was funded by the Ministry of Agriculture (Grant No. NAIK–MBK/M71411 and Grant No. NAIK–MBK/MSV002).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of Interest
Szemethy D. declares no conflict of interest. Mihalik B. declares no conflict of interest. Frank K. declares no conflict of interest. Nagy T. declares no conflict of interest. Újváry D. declares no conflict of interest. Kusza Sz. declares no conflict of interest. Szemethy L. declares no conflict of interest. Barta E. declares no conflict of interest. Stéger V. declares no conflict of interest.
Ethical Approval
All applicable international, national and/or institutional guidelines for the care and use of animals were followed.
Informed Consent
Not applicable.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Szemethy, D., Mihalik, B., Frank, K. et al. Development of Wild Boar Species-Specific DNA Markers for a Potential Quality Control and Traceability Method in Meat Products. Food Anal. Methods 14, 18–27 (2021). https://doi.org/10.1007/s12161-020-01840-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12161-020-01840-1