Abstract
The presence of pork content in food products can be a serious issue over an adulteration of food, particularly for consumers who pay attention to halal certification or other religious beliefs concerning the consumption of pork. Due to this fact, finding a method for the detection of pork content in food products that is sensitive, fast, and accurate is of interest. In this study, an electrochemical DNA biosensor with gold nanoparticle–DNA probe bioconjugates was developed to detect Sus scrofa mitochondrial DNA (mtDNA) using a gold-modified screen-printed carbon electrode (SPCE-Gold). The bioconjugates were formed by attaching the DNA probe with gold nanoparticles (AuNPs). Detection was carried out based on the optimum conditions resulting from the Box–Behnken experimental design that were 40 μL of 153 μg/mL bioconjugates; bioconjugate immobilization time, 20 min; and DNA hybridization time, 60 min. The hybridization of target synthetic DNA to the DNA probe was characterized by voltammetry based on the methylene blue indicator signal at a potential of about − 0.35 V. The results showed that the detection limit was 0.58 μg/mL and the recovery was 101.74%. mtDNA samples from various raw and processed meat were isolated and cut with restriction enzyme Sal1. Hybridization of mtDNA samples to the gold nanoparticle–DNA probe bioconjugates anchored to the SPCE-Gold surface was characterized based on the current response generated by methylene blue as an indicator. The result shows that there is an increasing trend in the response of raw and processed meat samples containing pork DNA. The development of an electrochemical DNA biosensor, based on gold nanoparticle–DNA probe bioconjugates, has not been reported previously. This biosensor is selective towards 10% of the pork DNA content in the mixture. Therefore, this biosensor method can be used as an alternative method to distinguish samples containing pork, according to the detection limit.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The growing findings of falsification in the meat information of food products have been a great concern, especially among consumers with religious beliefs that prohibit the consumption of pork. This concern has led to the importance of the development of an analytical method that would be able to identify different meat types in food products. Presently, there are various analytical methods available including the microscopic methods (Balizs et al. 2011), lipid-based methods with analysis of TAG and 2-MAG (Szabo et al. 2007), Fourier transform infrared (FTIR) spectroscopy (Rohman et al. 2011), protein-based methods (ELISA) (Giovannacci et al. 2004), and DNA-based methods (polymerase chain reaction/PCR) (Ali et al. 2012; Erwanto et al. 2011; Maryam et al. 2016; Ahmed et al. 2017). DNA-based analysis methods are advantageous given the stability of DNA reaction against heat, allowing the analysis of processed food products under high temperature. In addition, DNA extraction can be performed from all types of tissues because DNA is always present in each type of cell in the tissue (Aida et al. 2005).
Mitochondria are organelles that are abundant in cells and are the only organelles that have DNA. The sequence of mitochondrial DNA (mtDNA) of a species has a characteristic that can be distinguished from other species. The DNA sequence of the cytochrome b gene (cyt b) and the D-loop region are the most varied parts of mtDNA, so they can be used to differentiate species (Nguyen et al. 2016). Analysis of specific mtDNA sequences by the polymerase chain restriction-fragment length polymorphism (PCR-RFLP) method is the most commonly used method for identifying the presence of pork in food samples. The high sensitivity of the PCR-RFLP assay for the detection of pork content in commercial products is achieved due to the reduction of amplicon length (Bielikova et al. 2010; Ali et al. 2011). PCR-RFLP of a short fragment of degraded mtDNA in processed food is an option that can be applied (Ali et al. 2012). Analysis of mtDNA using oligonucleotide primers in the D-loop region and cyt b gene has been used to identify pig DNA in meat samples and commercially processed meat from the market, and other food products (Ali et al. 2012; Maryam et al. 2016). The real-time PCR method by using primers in the D-loop region and the cyt b gene is able to differentiate pig DNA from other species with the lowest concentration at 0.5% of pig DNA in a mixture of processed food products. High sensitivity is also obtained when both primers are used, with the detection limit as low as 5 pg/μL of pig DNA (Maryam et al. 2016). However, this method is less efficient given that it is time-consuming and it requires costly reagent and equipment, as well as involves high skills and expertise to be adopted. Several studies on the identification of meat types using mtDNA-based biosensor methods have been developed including gold nanoparticle (AuNP) colorimetric biosensors for identification of pork in meatballs (Erwanto et al. 2011), optical nanosensors for detection and quantification of DNA in meat mixtures (Ali et al. 2012b), and detection of pig DNA with a complex [Ru(bpy)2(PIP)] 2+(Halid et al. 2015). Recently, there has also been a determination of pork based on an optical fiber genosensor. These optical fiber biosensors rely on chemical-generated luminescence that has been shown to detect pork meat in the mixture in 1% quantity (Torelli et al. 2017).
Electrochemical DNA biosensor, on the other hand, is an advantageous method due to its simplicity, sensitivity, cost efficiency, and more importantly the portability of its devices, which enables practical application at any location (Lucarelli et al. 2004; Suryapratiwi et al. 2017). The use of bioconjugates in electrochemical biosensors has been widely developed to accelerate the detection process and increase the sensitivity. In addition, the use of bioconjugate with nanoparticles could enable the biosensors to be more selective and sensitive, and increase the ratios of signal versus blank (Emami et al. 2014; Hartati et al. 2019).
In this study, an electrochemical DNA biosensor method was developed using gold nanoparticle–cyt b DNA probe bioconjugate to detect pig mtDNA using the differential pulse voltammetry method. This biosensor is applied to pork samples, and other types of meat samples, including the processed meat sample. This method is expected to give an alternative method for detecting pig DNA in meat and food products.
Materials and Methods
Materials
Oligonucleotide DNA probe: 5′-/5ThioMC6-D/TACCGCCCTCGCAGCCGTAC-3′ and complementary DNA sequences: 5′-GTACGGCTGCGAGGGCGGTA-3′ were purchased from Integrated DNA Technologies Pte Ltd. (Singapore). DNeasy Mericon Food for DNA extraction and restriction enzyme Sal1 were from New England Biolabs (USA). N-Hydroxysuccinimide (NHS), 1-ethyl-3-(3-dimethylamino propyl) carbodiimide (EDC), gold(III) chloride hydrate (HAuCl4), 3-mercaptopropionic acid (MPA), 2′-deoxyadenosine 5′-triphosphate sodium salt (dATP), and sodium borohydride (NaBH4) were purchased from Sigma–Aldrich Ltd. (USA). Methylene blue (MB), sodium tetrahydridoborate (NaBH4), potassium ferric cyanide (K3[Fe(CN)6]), phosphate buffer, and PBS (phosphate-buffered saline containing 0.5 M NaCl) were purchased from Merck (Germany). Pork, beef, and chicken meat were obtained from the local supermarket. The processed food was prepared in a form of meatball which contains a mixture of pork, beef, and chicken meat in various percentage and they were cooked in the laboratory. All other chemicals and reagents were of analytical grade.
Apparatus
Cyclic voltammetry (CV) and differential pulse voltammetry (DPV) measurements were conducted on a μAutolab Type III Potentiostat/Galvanostat with NOVA 1.10 software (Metrohm, Switzerland). Screen-printed carbon electrode (SPCE) was obtained from GSI Technologies (USA). Other apparatus include a pH meter (Mettler Toledo InLab pH combination polymer electrodes), microcentrifuge (Thermo Scientific MicroCL 17R, USA), BDA digital compact gel documentation system, multi-mode reader (Tecan Infinite M200 PRO, Switzerland), UV/Vis spectrophotometer (Thermo Scientific, USA), and scanning electron microscope (SEM) (JEOL-JSM-6360LA, USA).
Preparation of AuNP–DNA Probe Bioconjugates
Colloidal AuNP was prepared based on sodium borohydride reduction (Wyantuti et al. 2015). The DNA-functionalized gold nanoparticle was done according to the following procedure (Yu et al. 2012): the 0.1 mM colloidal AuNP was incubated in 100 mM dATP in a mole ratio of 1:300, then the solution was allowed at room temperature for 15 min. Then, this solution was mixed with 10 mM PBS pH 7.4 (1:1 volume ratio with AuNPs). After brief vortexing, 0.1 mM DNA probe was introduced into AuNPs (30:1 mole ratio) by heating at 60 °C for 3 h to achieve equilibrium. The AuNP–dATP solution without DNA probe was also heated at 60 °C for 3 h as a control solution. Furthermore, both solutions were washed three times with 10 mM PBS pH 7.4, followed by centrifuging at 13,200 rpm for 20 min. The precipitate was dissolved in 100 μL of 10 mM PBS pH 7.4. The concentration of the bioconjugates obtained was 153.8 μg/mL. The AuNPs and AuNP–DNA probe bioconjugates were characterized by a UV-visible spectrophotometer. The UV-visible plasmon peak was normalized using R studio application software.
Modification of Screen-Printed Carbon Electrode Surface with Gold Nanoparticles
SPCE was pre-treated electrochemically by applying a potential of + 1.4 V for 60 min in 0.1 M phosphate buffer pH 7.0. The SPCE surface was dropped with 1.0 mM HAuCl4 and was electro-deposited by differential pulse voltammetry with the potential range from − 1.5 to 0.6 V at a scan rate equal to 0.1 V/s. Furthermore, the electrode is carefully rinsed with double distilled water and dried at room temperature. The SPCE-Gold electrodes then were characterized by cyclic voltammetry (CV) with a K3[Fe(CN)6] redox system in the potential range of − 0.6 to + 0.6 V and a scanning rate of 50 mV/s.
Immobilization of AuNP–DNA Probe Bioconjugates on SPCE-Gold Surface
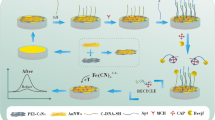
The SPCE-Gold was added with a linker, consisting of MPA, EDC, and NHS. Firstly, they were incubated in 0.1 M MPA solution for 1 h, then rinsed with 75% ethanol, and rinsed again with double distilled water. Furthermore, it was dropped with a 20-μL solution containing 0.1 M EDC and 0.1 M NHS with a ratio of 1:1, and incubated for 1 h at room temperature. The SPCE-Gold–linker electrode then was rinsed with a 0.05 M phosphate buffer pH 7.0. Furthermore, 40 μL of bioconjugates was dropped onto the SPCE-Gold–linker surface and incubated for 20 min. After the immobilization process was complete, the SPCE-Gold bioconjugate was rinsed with a 0.05 M phosphate buffer solution pH 7.0.
Optimization of Experimental Conditions Using the Box–Behnken Experiment Design
The optimization of the experiment was based on an experimental design using Box–Behnken analysis. Factors that influenced the experiment were optimized using the Minitab 18 program, which is bioconjugate volume/concentration, immobilization time of DNA probe–bioconjugates, and hybridization time between DNA probe and target DNA. Each factor was designed with three different levels which are the highest, the middle, and the lowest values, based on the literature and previous analysis (unpublished). The three experimental factors gave 15 times independent experiments. The Box–Behnken design was set to two replication blocks for three repetitions; therefore, the number of total experiments is 45 times. The interactions that resulted in the impact of reduced response were the bioconjugate immobilization time and the probe–target DNA hybridization time. Those interactions can be observed as response surface methods. The low area on the 3D chart indicates a low desirability value, while the high area indicates the high desirability value. The desirability function is a popular technique for the simultaneous determination of the optimum setting of input variables, so that it can determine the optimum level of performance for one or more responses (Islam et al. 2009).
Determination of Biosensor Response to Targeted DNA
SPCE-Gold bioconjugate was dropped with 20 μL of various concentrations of complementary DNA (0–30 μg/mL) in 0.05 M PBS pH 7.4, and incubated for 1 h. Then, the electrode was rinsed with 0.05 M phosphate buffer pH 7.0, dipped in methylene blue (MB) solution (12.5 μM MB, 0.05 M phosphate buffer pH 7.0 and 5.0 mM KCl) for 5 min, and then rinsed with 0.05 M PBS pH 7.0. The biosensor response was measured using the differential pulse voltammetry at the potential of − 0.45 V to + 0.1 V.
Extraction of DNA from the Samples
Approximately 200 mg of mashed samples (pork, beef, and chicken meat, and a mixture of processed meat) was weighed and placed into a 1.5-mL microtube. Then, the total DNA was isolated following the procedures in the DNeasy Mericon Food. The isolated DNA was then analyzed by electrophoresis on 1% agarose gel and quantified using a UV spectrophotometer.
Cutting DNA with Restriction Enzymes
Isolated DNA was cut with Sal1 restriction enzyme following the procedure. The reaction mixture contains 1 μg of DNA, 1 μL (20 units/μL) of Sal1 enzymes, 5 μL of 10× restriction buffer, and nuclease-free water to the total volume of 50 μL. The reaction mixture was incubated at 37 °C for 2 h, followed by electrophoresis analysis on 1% agarose gel and quantified by using a UV spectrophotometer.
Determination of DNA Biosensor Response on DNA samples
The 20 μL of DNA sample that had been restricted (~ 20 μg/mL) was diluted with 0.05 M phosphate buffer pH 7.0 in a total volume of 50 μL. DNA samples were denatured at 90 °C for 1 min, and immediately, 20 μL of the sample was dropped on SPCE-Gold bioconjugates and incubated for 1 h. The biosensor was then rinsed with 0.05 M phosphate buffer pH 7.0 and dipped in MB as mentioned in the “Determination of Biosensor Response to Targeted DNA” section. The biosensor response was measured using the differential pulse voltammetry at the potential of − 0.45 V to + 0.1 V.
Results and Discussion
AuNPs and AuNP–DNA Probe Bioconjugates
Figure 1 depicts the normalized spectra UV-visible absorption of AuNPs and AuNP–DNA probe bioconjugates with the same dilution (ten times). Absorption peaks were found at a wavelength of 529 nm for AuNPs and an additional peak at 260 nm for AuNP–DNA probe bioconjugates.
The color intensity of the AuNPs came from a photophysical response, which could be measured using a visible spectrophotometer at a wavelength of around 528 nm. The AuNP has a diameter of about 40 nm if it absorbs at this wavelength (Huang and El-Sayed 2010; Catherine and Olivier 2017). The plasmon peak of AuNP bioconjugates still appeared at about 528 nm, and the plasmon peak at 260 nm was the peak of the DNA probes that has been bound to the AuNPs. The higher intensity of DNA was due to the 30:1 mole ratio of DNA with AuNPs in bioconjugates.
The process of forming the AuNP–DNA probe bioconjugates involved the protection of AuNPs from salt-induced aggregation by adsorption of deoxyadenosine triphosphate (dATP) due to the repulsion of charges and steric obstruction between particles. dATP was chosen against four other mononucleotides due to its higher salt tolerance. In addition, dATP was also used as a spacer to reduce the number of DNA probes that will be bounded to the AuNP surface. If the AuNP was too dense, due to DNA probes, it would reduce the chances of the complementary DNA to hybridize.
Electrode Modification and Bioconjugate Immobilization
SEM image of SPCE and cyclic voltammograms of ferric cyanide on SPCE before and after gold modification are shown in Fig. 2.
(a1) SEM image of SPCE bare, (a2) SPCE-Gold, and (a3) SPCE-Gold bioconjugates. (b) Cyclic voltammogram as a result of SPCE characterization before and after modification with AuNP using 10 mM ferric cyanide (K3[Fe(CN)6]) in 0.1 M KCl with a scanning rate of 50 mV/s in the potential range of − 0.6 to + 0.6 V
Figure 2a1 shows the SEM image of bare SPCE at × 500,000 magnification. In Fig. 2a2, SEM image of SPCE-Gold shows that there is the attachment of reduced gold from HAuCl4. It can be seen that the carbon surface was covered by Au nanostructure. Figure 2a3 shows that the carbon surface was more covered after the DNA immobilization. Figure 2b reflects that the peak current of ferric cyanide increases after SPCE was modified with gold. The rising of redox peak current was associated with an increase of electrochemical activity with increasing electrode active area.
The immobilization process of AuNP–DNA probe bioconjugates on the SPCE surface involves MPA as a linker. The thiol (–SH) group of MPA was used as a binding site for covalent bond with metal nanoparticles, while the carboxylic group (–COOH) was used to react covalently with phosphate groups in DNA to achieve effective immobilization. The carboxylic group of MPA was activated by EDC and NHS so that a succinimide ester is formed which was used to immobilize the bioconjugates on SPCE surfaces.
DNA Biosensor Responses
Duplex hybridization formation will occur directly through the diffusion of complementary DNA to the DNA probe, which was bound to the AuNP as bioconjugates. One of the factors influencing hybridization process is the density of ssDNA attached to AuNP. The hybridization process will decrease in compliance with the increase of ssDNA density by AuNP, because of the electrostatic repulsion produced by the negatively charged phosphate group from ssDNA. This indicates the importance of controlling the density of ssDNA which is attached to the surface of the AuNP (Farjami et al. 2010). Therefore, the use of dATP as a spacer provides a more flexible and free space for complementary DNA, which will increase the initiation of duplex formation and in turn result in an increased hybridization rate.
The DNA biosensor response to SPCE-gold bioconjugate–target DNA was observed based on the MB indicator signal at − 0.34 V to − 0.37 V. The increase of peak current and a potential positive shift occur in MB–dsDNA. This potential shift is typical of MB intercalation with dsDNA. The increase of peak current correlated to the number of MB molecules that have been intercalated to the double helix structure of the dsDNA. Although all DNA probes have hybridized to form a duplex, the voltammetric signal could still be observed, as MB would also act as an intercalator. Besides MB interacts with dsDNA by intercalation between G–C base pairs, and positively charged MB molecules generally are bound with the phosphate groups of DNA electrostatically. The electrochemical signal of MB bound to dsDNA is determined by the distance of transferring electrons from the MB molecule to the electrode, which was intercalated or electrostatically bound at dsDNA (Kira et al. 2015). The transfer of electrons through dsDNA was limited by a sequence length of up to about 18–20 nucleotides. Only MB molecules were located closer to the surface of the electrode, which could be involved in the electron transfer, and were responsible for the electrochemical responses (Tavallaie et al. 2014).
The SPCE modification scheme, bioconjugate immobilization, and hybridization of probe–target DNA are shown in Fig. 3.
Optimization Data Using the Box–Behnken Experiment Design
The interactions between factors that influenced the experiments, that are bioconjugate volume/concentration, immobilization time of DNA probe bioconjugates, and hybridization time of DNA probe–target DNA, can be observed as a response surface method shown in Fig. 4. The low area on the 3D chart indicates a low desirability value, while the high area indicates a high desirability value. The resulting desirability value between 0 and 1 indicates the closeness of the response to the ideal value. In this study, a desirability value of 0.9354 was obtained. As the desirability value produced was near to 1, it indicates that the response in this experiment was close to the ideal value.
Based on the ANOVA results, the optimum experimental conditions obtained through the Box–Behnken experiment design were 40 μL of 153 μg/mL bioconjugates, 20 min of bioconjugate immobilization time, and 60 min of complementary DNA hybridization time.
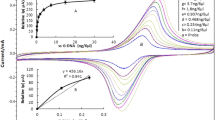
Calibration Graph of Various Targeted DNA Concentrations
The calibration graph of the range of target DNA concentrations from 0.1 to 5.0 μg/mL as a dynamic range of this sensor could be observed in Fig. 5, and the equation of the line is Y = 0.2223 X + 1.6249 with linearity (R2) of 0.9853. The measurement of the peak current up to 5.0 μg/mL of target DNA shows the R value that is lower than 0.9. Therefore, a line equation with a linear concentration range of 0.1–5.0 μg/mL was used to calculate the limit of detection.
Figure 5 reflects that in more concentrated target DNA, the number of MB molecules intercalated to dsDNA was increased, allowing for more MB molecules to be located closer to the electrode surface, so that electron transfers are more facilitated and higher peak current could be produced.
Limit of detection (LOD) was determined using the equation Y = Yb + 3Sb, Yb is the intercept of calibration curve (a) and Sb is the SX/Y which estimates the random errors in the y-direction (Miller and Miller 2010). The LOD value was 0.58 μg/mL, limit of quantitation (LOQ) value was 1.94 μg/mL, and recovery was 101.74%. A previous research using an electrochemical DNA biosensor using graphene as modifier of screen-printed carbon electrode has found LOD at 1.76 μg/mL (Hartati et al. 2019), and swine DNA in processed beef LOD was found to be at 4.0 μg/mL and 6.0 μg/mL, respectively, using gold nanoparticles as colorimetric sensors (Ali et al. 2011b, b). The LOD of dot blot for pork sample as a sensitive and selective test is positive at 2.0 ng/μL using an optical fiber chemiluminescence biosensor (Torelli et al. 2017). The detection limit obtained in this electrochemical method can be compared with previous biosensor methods and can be used as an alternative and a simple method to distinguish raw meat samples and processed meat.
Analysis of Sample
DNA of pork, beef, and chicken was isolated using DNeasy Mericon Food. Figure 6 depicts the electrophoregram of isolated DNA on 1% agarose gel. The quality and quantity of DNA were measured using a UV spectrophotometer (Table 1).
The isolated DNA was cut with Sal1 restriction enzyme and was characterized by agarose electrophoresis (data was not shown). The Sal1 enzyme recognizes and cut at one site on mtDNA to produce ~ 16,731-bp linear DNA in size. Linear mtDNA was used to measure the response and selectivity of the electrochemical DNA biosensor method that was developed.
Biosensor Response of Pork mtDNA from Meat and Meatball Samples
The DNA biosensor response of mtDNA which was extracted directly from pork, chicken, and beef raw meat, and samples of the four mixed processed meat (1) 10% Pork:25% chicken:65% beef, (2) 25% Pork:25% Chicken:50% Beef, (3) 50% Pork:50% Beef, and (4) 50% Pork:50% Chicken were determined. There was a decrease in the average of peak current when compared between pre- and post-processed meat of each component as shown in Table 2; this might be due to DNA damage in the cooking process. The peak current decreased about 15 to 20%. The voltammograms of processed meat are depicted in Fig. 7.
Figure 7 shows that the peak current response of 100% negative sample (chicken DNA and beef DNA only) was almost of the same height with AuNP–DNA probe bioconjugates; this indicated that mtDNA of chicken and beef could not hybridized to DNA probe. Moreover, the peak current of mix processed meat showed that the more percentage of pork content, the higher the peak current, and the highest peak current is in 100% pork. Voltammogram in Fig. 7 was chosen from three results of the measurement. Furthermore, it can be seen that there is a difference in the height of the peak current for the same percentage of pork. The peak current of 6 and 7 contains 50% pork, but mixed with different meat. The difference in peak current might be influenced by percentage of homology between DNA probe and mtDNA of chicken and beef. However, the calculation of the average peak current showed the close values (data not shown). The measurement was thought to be reproducible. In addition, the 10% pork sample (4) which consists of approximately 0.8 μg/mL of the pig DNA showed the difference of peak current about 16% compared to 100% chicken and beef. This data indicated that the biosensor can clearly detect the pork DNA even though the pork content is 10% (10:100 ratio). The comparison of performance between the current biosensor and others can be seen in Table 3.
Although this method has not been tested to a mixture with 1% pork which was previously investigated by using an optical chemiluminescence genosensor (Torelli et al. 2017), this method can still be an alternative to distinguish between raw meat, processed meat, and sample with 10% pork content.
Conclusion
Electrochemical DNA biosensors based on AuNP–DNA probe bioconjugates are capable of distinguishing the DNA samples from pork, chicken, and beef. The developed electrochemical DNA biosensor was selective in detecting pig mtDNA, as evidenced by the difference in peak currents generated from mixture of processed meat samples. The detection limit obtained with this method can be compared with previous biosensor methods.
References
Ahmed SA, Pin LL, Raabe CA (2017) A combined rapid DNA extraction and multiplex PCR for the detection of porcine DNA in raw and processed food. Contemporary Issues and Development in the Global Halal Industry, 201-208
Aida AA, Man YBC, Wong CMVL, Raha AR, Son R (2005) Analysis of raw meats and fats of pigs using polymerase chain reaction for Halal authentication. Meat Sci 69(1):47–52
Ali ME, Hashim U, Mustafa S, Che Man YB (2011) Swine-specific PCR-RFLP assay targeting mitochondrial cytochrome B gene for semiquantitative detection of pork in commercial meat products. Food Anal Methods 5:613–623
Ali ME, Hashim U, Mustafa S, Che Man YB (2011b) Nanoparticle sensor for label free detection of swine DNA in mixed biological samples. Nanotechnology 22:195503. https://doi.org/10.1088/0957-4484/22/19/195503
Ali ME, Mustafa S, Hashim U, Che Man YB, Foo KL (2012) Nanobioprobe for the determination of pork adulteration in burger formulations. J Nanomater 2012:1–7
Ali ME, Mustafa S, Che Man YB, Islam KN (2012b) Gold nanoparticle sensor for the visual detection of pork adulteration in meatball formulation. J Nanomater 2012:1–7
Balizs G, Weise C, Rozycki C, Opialla T, Sawada S, Zagon J, Lampen A (2011) Determination of osteocalcin in meat and bone meal of bovine and porcine origin using matrix-assisted laser desorption ionization/time-of-flight mass spectrometry and high-resolution hybrid mass spectrometry. Anal Chim Acta 693(1–2):89–99
Bielikova M, Pangallo D, Turna J (2010) Polymerase chain reaction - restriction fragment length polymorphism (PCR-RFLP) as a molecular discrimination tool for raw and heat-treated game and domestic animal meats. J Food Nutr Res 49:134–139
Catherine L, Olivier P (2017) Gold nanoparticles for physics, chemistry and biology. 2nd ed. World Scientific, London
Emami M, Shamsipur M, Irajirad R (2014) A breast cancer biomarker based on antiHER2–iron oxide nanoparticle bioconjugates. Analyst 139:2858–2866
Erwanto Y, Abidin MZ, Rohman A, Sismindari S (2011) PCR-RFLP using BseDI enzyme for pork authentication in sausage and nugget products. Media Peternakan 34(1):14–18
Farjami E, Clima L, Gothelf KV, Ferapontova EE (2010) DNA interactions with a methylene blue redox indicator depend on the DNA length and are sequence specific. Analyst. 135(6):1443–1448
Giovannacci I, Guizard C, Carlier M, Duval V, Martin JL, Demeulemester C (2004) Species identification of meat products by ELISA. Int J Food Sci Technol 39(8):863–867
Halid NIA, Hasbullah SA, Ahmad H, Heng LY, Karim NHA, Harun SN (2015) Electrochemical DNA biosensor for detection of porcine oligonucleotides using ruthenium (II). Conference Proceeding
Hartati YW, Setiawati TA, Sofyatin T, Fitrilawati, Anggraeni A, Gaffar S (2019) Electrochemical DNA biosensor for pork (Sus scrofa) determination using screen printed carbon-reduced graphene oxide electrodes, Scienceasia (Under review)
Huang X, El-Sayed MA (2010) Gold nanoparticles: optical properties and implementations in cancer diagnosis and photothermal therapy. J Adv Res 1(1):13–28
Islam MA, Sakkas V, Albanis TA (2009) Application of statistical design of experiment with desirability function for the removal of organophosphorus pesticide from aqueous solution by low-cost material. J Hazard Mater 170(1):230–238
Kira A, Matsuo K, Nakajima S (2015) DNA hybridization activity of single-stranded DNA-conjugated gold nanoparticles used as probes for DNA detection. Jpn J Appl Phys 55 (2S)
Lucarelli F, Marrazza G, Turner APF, Mascini M (2004) Carbon and gold electrodes as electrochemical transducers for DNA hybridisation sensors. Biosens Bioelectron 19:515–530
Maryam S, Sismindari S, Raharjo TJ, Sudjadi, Rohman A (2016) Determination of porcine contamination in laboratory prepared dendeng using mitochondrial D-Loop686 and cyt b gene primers by real time polymerase chain reaction. Int J Food Prop 19(1):187–195
Miller JN, Miller JC (2010) Statistics and chemometrics for analytical chemistry, Sixth Edition. Pearson Education Limited, England
Nguyen HD, Bui TA, Nguyen PT, Kim OTP, Vo TTB (2016) The complete mitochondrial genome sequence of the indigenous I pig (Sus scrofa) in Vietnam. Asian Australas J Anim Sci 30(7):930–937
Rohman A, Sismindari, Erwanto Y, Che Man YB (2011) Analysis of pork adulteration in beef meatball using Fourier transform infrared (FTIR) spectroscopy. Meat Sci 88(1):91–95
Suryapratiwi WN, Paat VI, Gaffar S Hartati YW (2017) DNA biosensor for detection of Salmonella typhi from blood sample of typhoid fever patient using gold electrode modified by self-assembled monolayers of thiols, AIP Conference Proceedings
Szabo A, Febel H, Sugar L Romvari R (2007) Fatty acid regiodistribution analysis of divergent animal triacylglycerol samples - a possible approach for species differentiation. Journal of Food Lipids complex as intercalator label redox AIP Conference Proceedings 313(August), 313–316.
Tavallaie R, Darwish N, Gebala M, Hibbert DB, Gooding JJ (2014) The effect of interfacial design on the electrochemical detection of DNA and microRNA using methylene blue at low-density DNA films. ChemElectroChem. 1(1):165–171
Torelli E, Manzano M, Marks RS (2017) Chemiluminescent optical fibregenosensor for porcine meat detection. Sensors Actuators B Chem 247:868–874
Wyantuti S, Hartati YW, Firdaus ML, Panatarani C, Tjokronegoro R (2015) Fabrication of gold nanoparticles-modified glassy carbon electrode and its application for voltammetric detection of Cr (III). Int J Sci Technol Res 4:135–139
Yu HLL, Montesa CM, Rojas NRL, Enriquez EP (2012) Nucleic-acid based lateral flow strip biosensor via competitive binding for possible dengue detection. Biosens Bioelectron 3:128. https://doi.org/10.4172/2155-6210.1000128
Acknowledgments
The authors would like to thank Research Grant of Padjadjaran University (no. 3771/UN6.D/LT/2018) for financial support. Our sincere thanks also go to Dr. Rani Maharani for proofreading the manuscript and to Dr. Ari Hardianto for normalizing the UV-visible data by using R studio software.
Funding
This research was financially supported by Universitas Padjadjaran Research Grant of Scheme Riset Kompetensi Dosen Universitas Padjadjaran to Dr. Anni Anggraeni, M.Si (no. 3771/UN6.D/LT/2018.
Author information
Authors and Affiliations
Contributions
Y.W-H., A.A-S, M.-A performed the experiments; A.-A and G.-S conceived and designed the experiments; Y.W-H and G.-S analyzed the data and wrote the paper.
Corresponding author
Ethics declarations
Ethical Approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Informed consent
Not applicable.
Conflict of Interest
Yeni Wahyuni Hartati declares that she has no conflict of interest. Anis Amiliya Suryani declares that she has no conflict of interest. Mila Agustina declares that she has no conflict of interest. Shabarni Gaffar declares that she has no conflict of interest. Anni Anggraeni declares that she has no conflict of interest.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Hartati, Y.W., Suryani, A.A., Agustina, M. et al. A Gold Nanoparticle–DNA Bioconjugate–Based Electrochemical Biosensor for Detection of Sus scrofa mtDNA in Raw and Processed Meat. Food Anal. Methods 12, 2591–2600 (2019). https://doi.org/10.1007/s12161-019-01593-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12161-019-01593-6