Abstract
Two studies examined genetic and environmental influences on traits proposed by the revised Reinforcement Sensitivity Theory (rRST) of personality. Both quantitative and molecular behavioral genetic methods were applied considering the effects of COMT, DRD2, HTR1A and TPH2 single nucleotide polymorphisms (SNPs). Study one included 274 monozygotic and 154 dizygotic twins for the quantitative behavioral study; and in study two there were 431 twins for the molecular genetic study. The Reinforcement Sensitivity Questionnaire was used to assess basic personality traits defined by the rRST. Univariate biometric modeling suggested that genetic influences accounted for 34–44% of variance of Behavioral Approach System (BAS), Behavioral Inhibition System (BIS) and Fight-Fligh-Freeze System. Molecular genetic analyses proposed the significant main effect of COMT SNP on the BAS and TPH2 SNP on the BIS, and pointed out epistatic effects of COMT x DRD2 on BAS and HTR1A x TPH2 on Fight. Results demonstrated substantial heritability for all rRST constructs, as well as for differences in the molecular genetic basis of both approach-related and avoidance-related dimensions.
Similar content being viewed by others
Introduction
As one of the most prominent conceptual frameworks in the field of “personality neuroscience”, the Reinforcement Sensitivity Theory (RST) emphasizes the impact of neurophysiological factors on individual differences in human behavior (Corr & McNaughton, 2012; Gray & McNaughton, 2000) – it also points to the role of genetic factors in shaping the behavioral patterns in reaction (dis)incentives of various kinds. The original RST (Gray, 1982) assumed the existence of three major neurological systems: The Behavioral Approach System (BAS) was regarded as the basis of reactions to conditioned appetitive stimuli; the Behavioral Inhibition System (BIS) was said to underlie reactions to conditioned aversive stimuli; and the Fight/Flight system (FFS) was thought responsible for reactions to unconditioned aversive stimuli (Gray, 1982). Within the revised RST (rRST; Gray & McNaughton, 2000): BIS has been redefined as the basis for the processing of conflicting stimuli; BAS has become responsible for reactions to all appetitive stimuli; Fight/Flight/Freeze system (FFFS) underlies defensive reactions regardless of the conditionality of the stimulus. The BIS corresponds to anxiety and is more concerned with anticipated or potential (instead of actual) threats. In contrast, present threats can activate the fear-related FFFS, which in turn can trigger reactions such as confrontation, escape, or a temporary halt in behavioral reactions, which correspond to aggression, phobias and panic reactions, respectively (e.g., Gray & McNaughton, 2000). Despite the reformulation, the content of the BIS and BAS dimensions, which were most used in previous research, has not been substantially altered. Namely, BAS in rRST remains a system associated with impulsivity, and BIS system associated with anxiety.
In this study, the RSQ (Smederevac et al., 2014) was used, which was previously validated in several studies, focusing on various issues: distinguishing between anxiety and fear (Mihić et al., 2015), experimental approach to social anxiety (Ranđelović et al., 2018), prisoners’ personality prototypes (Mitrović et al., 2014), and psychological response to the Covid 19 pandemic (Sadiković et al., 2020).
Certain conceptual similarities between the dimensions, regardless of questionnaires (Corr & Cooper, 2016; Jackson, 2009; Reuter et al., 2015; Smederevac et al., 2014) lead to their non-orthogonal relationship, resulting in medium correlations between BAS and Fight, as well as between BIS, Flight and Freeze, which correspond to a broader approach-avoidance strategies (Corr & McNaughton, 2012).
So far, no quantitative behavioral genetic study has assessed rRST personality factors, and molecular genetic studies have seldom been conducted (Reuter et al., 2006; Reuter et al., 2015) Only one longitudinal twin study (Takahashi et al., 2007) has addressed stability and change of genetic and environmental factors underlining BIS and BAS using the BIS/BAS scale (Carver & White, 1994), related to original RST. Results showed that the phenotypic manifestations of BIS and BAS are affected primarily by non-shared environmental factors (proportion of explained variance spanning 66–67% for BIS and 65–72% for BAS), and to a lesser extent by additive genetic factors (33–34% for BIS and 28–35% for BAS). Although this study used the BIS/BAS scales, their content has not been substantially changed in the rRST and these results can be considered highly relevant, but have not been replicated so far. However, several behavioral genetic studies have examined personality traits that are similar to the RST/rRST constructs, by name or by content. In these cases, additive genetic influences were estimated at approximately: 38–45% for impulsivity (e.g., Bezdjian et al., 2011) which represents the basic content of BAS; 47% for direct assault, an aspect of aggressiveness (e.g., Coccaro et al., 1993) related to the Fight; 40–55% for anxiety (e.g., Gagne et al., 2016) which is the basic content of the BIS; 0–71% for fears and specific phobias (e.g., Houtem et al., 2013), depending on particular types; 35–45% for fear conditioning (e.g., Hettema et al., 2003); and 43% for panic disorder (e.g., Bienvenu et al., 2011) which is associated with Freeze.
Molecular genetic studies on RST constructs have demonstrated relations of different gene polymorphisms to personality traits (Montag et al., 2010; Pecina et al., 2013; Reuter et al., 2006; Smillie et al., 2010; Tunbridge et al., 2019), producing inconsistent results (Johnson et al., 2016). In the present study, DRD2 (rs1800497), COMT (rs4680), HTR1A (rs6295) and TPH2 (rs4570625) genes, most commonly associated with responses to rewards and punishments, as the core concepts of rRST, were selected. DRD2 and COMT genes are predominantly expressed in the brain tissue of cortex, as receptors (e.g., Pecina et al., 2013) and enzymes (e.g., Chen et al., 2011) involved in dopaminergic pathways, regulating dopamine concentration (Smillie et al., 2010; Tsuchimine et al., 2012) and degradation (DeYoung et al., 2011; Tunbridge et al., 2019). On the other hand, HTR1A and TPH2 genes are involved in serotonergic system, and expressed in the brain tissue, regulating serotonin biosynthesis, as serotonin enzymes (e.g., Aoki et al., 2010; Lehto et al., 2015), and transmission (e.g., Benko et al., 2010), as receptors of serotonin (e.g., Koller et al., 2006).
One of the key genes that seems to contribute to the variation in the reward and emotional processing is a D2 receptor gene (DRD2), member of D2 family, located on chromosome 11q22-q23, and highly expressed in striatum and prefrontal cortex (e.g., Pecina et al., 2013). Reward is governed by the mesolimbic reward system that involves dopaminergic pathways, DRD2 variation explained only 2.5% of the variance in Extraversion (Smillie et al., 2010), and that exclusively male carriers of DRD2/ANKK1 A1 allele scored higher in reward dependence, related to BAS. More revealing interactions of the DRD2 with other genes, such as COMT, have shown more convincing relations with impulsiveness, reward responsiveness and related behaviors (Reuter et al., 2006). Aluja et al. (2019) showed that DRD2 gene, along with other genes on dopaminergic pathway, explain around 14.5% of Sensation Seeking.
The catechol-O-methyltransferase (COMT) gene, located on chromosome 22q11-q12, has a substantial role in the enzymatic degradation of adrenaline, noradrenaline, and dopamine (Akil et al., 2003; Chen et al., 2011). Although the COMT gene contains numerous polymorphisms, one of the most studied is Val 158Met (rs4680), which encodes three possible genotypes: Val/Val, Met/Val, and Met/Met. Previous studies (e.g., Chen et al., 2011) suggested that Met carriers showed lower activity of COMT enzyme for around 30–35% in prefrontal cortex, resulting in increased dopamine concentration (e.g., Demetrovics et al., 2010).
Kotyuk et al. (2015) showed that carriers of the Val/Val genotype have lower mean scores on neuroticism, related to BIS, and higher mean scores on agreeableness, related to Fight, as compared with carriers of the Met/Met or Met/Val genotypes. On the other hand, some other studies showed that Met carriers have higher anxiety (Valeeva et al., 2020) and lower extraversion (Hoth et al., 2006), but only in female sample, whereas results were opposite in men sample: Val carriers showed higher negative emotionality (Chen et al., 2011) and lower extraversion (Demetrovics et al., 2010) which correspond to BIS and BAS, respectively.
Despite inconsistent results, studies conducted so far provide sufficient evidence for the view that COMT Val-allele is usually associated with an advantage in emotional and the Met-allele in cognitive processing (for further details see Mier et al., 2010). Relying on these findings, Goldman et al. (2005) postulated the Warrior–Worrier hypothesis, which posits a cognitive/emotional trade-off, connecting Warrior strategy with Val allele, and Worrier strategy with Met allele.
Within the RST framework, previous studies have shown that higher BAS scores, in particular on the ‘Drive’ and ‘Fun-seeking’ factors of the Carver and White (1994) BIS/BAS Scales, are associated with low COMT activity and high DRD2 receptor density, but also with high COMT activity and low DRD2 receptor density (Reuter et al., 2006). Reuter et al. postulated the COMT–DRD2 Equilibrium Model of Positive Emotionality, explaining these patterns of alleles which were associated with behavioral approach and PEM (Reuter et al., 2006). This COMT and DRD2 epistasis effect has been confirmed in studies of cognitive processes (e.g., Garcia-Garcia et al., 2011), while it has not been confirmed in a sample of depressed patients (Light et al., 2007).
The serotonergic system has an important role in the regulation of anxiety and depression (Whisman et al., 2011), as well as impulsivity and aggressive behavior (Nelson & Trainor, 2007). HTR1A receptor (5-HT1A) is the major autoreceptor on serotonergic raphe neurons, and its gene HTR1A is located on human chromosome 5q12.3. Several studies have demonstrated that G variant of the HTR1A polymorphism is associated with depression, suicidality, and PTSD (Goçi Uka et al., 2019; Lemonde et al., 2003), anxiety (Strobel et al., 2003), aggression (Witte et al., 2009) and general amygdala reactivity (e.g., Kalbitzer et al., 2009). Some studies suggested that G carriers are showing higher scores on Neuroticism (Schinka et al., 2004) and even impulsivity (e.g., Gray et al., 2018), but other studies did not report these connections (e.g., Kalbitzer et al., 2009; Koller et al., 2006).
Tryptophan hydroxylase-2 (TPH2) is the rate-limiting neuron-specific isoform catalyzing the rate-limiting step in biosynthesis of 5-HT in the brain, and its gene TPH2 is located on chromosome 12q21. This gene has an important role in regulation of the serotonergic system (Kautzky et al., 2019), aggressive behavior, cognitive control, and emotional regulation (Lehto et al., 2015; Waider et al., 2011). It is shown that certain allelic forms could promote lower anxiety, impulsivity, and aggressiveness (Laasa et al., 2017), but results are inconsistent (Gutknecht et al., 2007). Previous studies demonstrated the correlation between amygdala–hippocampus connectivity and BIS system in individuals homozygous for the TPH2 G allele (Hahn et al., 2013). Moreover, the T allele of TPH2 (rs4570625) is hypothesized to be associated with reduced serotonin levels. One meta-analysis (Lehto et al., 2015) showed that there are epistatic effects of HTR1A and TPH2 genes on Conscientiousness.
Current Study
This study relies on an approach of combining quantitative and molecular genetic design to delineate causal pathways underlying approach and avoidance strategies, arising from rRST (Corr & McNaughton, 2012).
Here we examine the behavioral genetic and molecular genetic bases of the rRST personality traits, measured by the RSQ (Smederevac et al., 2014) – these different analyses were performed on the almost same twin sample in two different studies. Namely, the only differences in the samples were caused by the missing a few genotypic data. Since there are phenotypic similarities between approach-related traits, such as BAS and Fight, as well as avoidance-related traits, such as BIS, Flight and Freeze (Corr & Cooper, 2016; Reuter et al., 2015; Smederevac et al., 2014), we have examined genetic and environmental factors that might provide a further explanation of their neurophysiologic and genetic foundations. Using a multivariate Cholesky decomposition twin model to partition covariation into genetic and environmental components, we examined the degree to which the genes and environment contribute to the overlap between the traits. We were particularly interested in determining the degree to which the genetic influences on traits would be entirely accounted for by common genetic factors or whether some proportion of the genetic influence would be specific. We hypothesized that rRST traits, in addition to common genetic, would have specific sources of genetic and environmental variance, which may facilitate empirical evidence for their hierarchical structure.
Next, we have examined the possible association between COMT rs4680, DRD2 rs1800497, HTR1A rs6295 and TPH2 rs4570625 gene polymorphisms and BIS, BAS, and FFFS. Despite the inconsistent results regarding the molecular genetic basis of personality traits (Johnson et al., 2016), we hypothesized that the dopamine related DRD2 and COMT genes would form the basis for the approach-related behavior, such as BAS and Fight, while HTR1A and TPH2 genes, as regulators of serotonin level would be important for both approach and avoidance strategies. Dopaminergic pathways are regulating approach behaviors through reward processing (e.g., Aarts et al., 2010) and incapability to learn from punishments (e.g., Whitmer & Gotlib, 2012), which are the distinctive features of BAS and Fight. Besides the main effects, we hypothesized the interaction of dopamine genes on BAS, which would be an extremely important replication of previous findings (Reuter et al., 2006).
On the other hand, increasing in serotonin transmission contributes cognitive flexibility and approaching behaviors, but decreasing could lead to higher sensibility to environments, stress, and avoidance behaviors (e.g., Goçi Uka et al., 2019; Kalbitzer et al., 2009). Therefore, the main effects of HTR1A and TPH2 on the avoidance strategies, covered by the BIS, Flight and Freeze, are expected.
We do not have specific hypotheses about the interaction of serotonin genes since previous research has mainly examined their common additive effects on traits related to emotional regulation (Pearson et al., 2014; Spagnolo et al., 2020). Namely, although previous findings indicate robust effects of HTR1A and TPH2 genes on anxiety and depressive responses (Alqahtani et al., 2020; Mandelli et al., 2012), which corresponds to BIS, their role in the regulation of behaviors associated with lack of control should not be neglected (Juhasz et al., 2009; Pearson et al., 2014). In other words, previous findings imply that examination of the role of serotonin genes should not be limited to avoidance strategies.
Method
Procedure and Sample
The Serbian Twin Registry has data on 1654 participants. The entire procedure for testing and collecting data is described elsewhere (Smederevac et al., 2019). Participation was voluntary and participants signed informed consent. The research was approved by the Institutional Ethical Committee (approval certificate: http://psihologija.ff.uns.ac.rs/etika/?odobreno=20111020000004_e1b8).
The participants with substantial amounts of missing data (e.g. missing responses to a more than a third of a questionnaire items) were removed from the sample before the analyses. Missing data were replaced using the full-information maximum likelihood procedure, as mentioned in Brown (2015) and implemented in Rosseel (2012).
This study covers both the quantitative behavioral genetic and molecular genetic aspects of the rRST dimensions in the same sample of adult twins. However, due to missing data, there are small differences in sample size in these two analyses. In the quantitative behavioral genetic analysis, participants were 274 monozygotic twins (60 males, 212 females), age ranged 17 to 60 (M = 26.04; SD = 8.44); and 154 dizygotic twins (55 males and 98 females) age ranged 17 to 48 (M = 22.91; SD = 6.05).
The initial sample in molecular genetic analyses consisted of 433 twins (25.4% males and 73.4% females) age ranged from 17 to 60 (M = 24.69, SD = 7.71). After excluding participants with inappropriate buccal swab, the final sample consisted of 431 twins for COMT and DRD2 genes, 423 for HTR1A and 390 for TPH2 gene. We considered it more appropriate to use all available data than to throw out useful data for the same number of subjects into all analyzed gene polymorphisms.
The determination of zygosity was conducted using a DNA buccal swab analysis. Buccal swabs were tested using short tandem repeat (STR) megaplex kits, either Investigator 24plex GO! (Qiagen®, Valencia, CA, USA). Kit detects 21 autosomal and two gender markers, Amelogenin and DYS391. Samples with partial profiles were interpreted if at least 10 loci had results. Both quantitative behavioral genetic and molecular genetic datasets are deposited at OSF (https://osf.io/csh23/).
Measures
Reinforcement Sensitivity Questionnaire (RSQ; Smederevac et al., 2014) is based on the Revised Reinforcement Sensitivity Theory (Gray & McNaughton, 2000). It contains 29 items on five scales: Behavioral inhibition system - BIS (7 items, α = .85), Behavioral activation system - BAS (6 items, α = .76), Fight, Flight and Freeze system (with 5 items each, α = .81, .69, .86, respectively). Items are rated on a four-point Likert scale (from 1 = completely disagree to 4 = completely agree).
Genotyping of DRD2, COMT, HTR1A and TPH2 Polymorphisms
DNA was extracted using standard DNA extraction techniques (QIAgen®, Hilden, Germany) as recommended by the manufacturer. The genotyping of the DRD2 gene (rs1800497), COMT gene (rs4680), TPH2 gene (rs4570625) and HTR1A (rs6295) was carried out using TaqMan assays (TaqMan SNP, Applied Biosystems®, Warrington, UK), as recommended by the manufacturer. The TaqMan SNP Genotyping Assays uses TaqMan 5′ -nuclease chemistry for amplifying and detecting specific polymorphisms in purified genomic DNA samples and takes advantage of minor groove-binding probes for superior allelic discrimination. The SNP Genotyping Assays contain a VIC-dye-labelled probe, FAM-dye-labelled probe with two target-specific primers. PCR was performed using 10 ng of genomic DNA together with 1 μl TaqMan Genotyping assay and 12.5 μl of genotyping master mix in final 25 μl reaction on a 96-well plate using ABI Prism 7500 Fast PCR device (Applied Biosystems®, Foster City, California, USA). DRD2, COMT, TPH2 and HTR1A alleles with the specific fluorescence curves were detected and analyzed using the 7500 System SDS program, integrated into the ABI Prism 7500 Fast PCR device.
The COMT rs4680 gene polymorphism was initially defined by 3 groups: 130 high activity homozygotes (Met/Met), 224 intermediate heterozygotes (Met/Val) and 77 low activity homozygotes (Val/Val) (Table 1). Val/Val and Met/Val genotypes were combined into a single group (Val+) with 301 participants, while Met/Met formed Met+ group with 130 participants. COMT rs4680 was in Hardy–Weinberg equilibrium (HWE), with no significant differences between observed and calculated genotype frequencies (χ2 = 0.83, df = 1, p > .05).
Initially, DRD2 rs1800497 gene polymorphism was defined in 3 groups: 13 were high activity homozygotes (A1), 120 were intermediate heterozygotes (A1/A2), and 298 were low activity homozygotes (A2) (Table 1). A1 homozygotes and A1/A2 heterozygotes were combined into a single group (A1 +). After integration, the number of A1+ and A2 (A1–) genotypes were 133 and 298, respectively. DRD2 rs1800497 was in HWE (χ2 = 2.79, df = 2, p > .05).
Among the study sample, 99 individuals did not carry the HTR1A rs6295 G allele (i.e., C/C genotype). However, 230 and 94 subjects had one (i.e., C/G genotype) or two copies (i.e., G/G genotype) of the HTR1A rs6295 G allele, respectively. The genotypes with G allele were integrated in a single CG group (C/G and G/G genotype) with 324 participants, while the CC (C/C genotype) group consisted of 99 participants (Table 1). The genotype distribution of HTR1A did not deviate significantly from HWE (χ2 = 3.25, df = 2, p > .05).
TPH2 rs4570625 gene polymorphism resulted in 3 genotypes: GG (243 participants), GT (146 participants) and TT (only 1 participant). The genotypes with T allele were grouped by T carriers (T/G and T/T genotypes) and resulted with 147 participants in GT group (Table 1). Since TPH2 genotype frequencies were not in the HWE equilibrium (χ2 = 17.318, df = 2, p < .05), we have tested TPH2 allelic configurations, which are in the HWE equilibrium (χ2 = 1.98, df = 1, p > .05).
All analyzes were performed at the allele level, so that all subjects with risk alleles were classified into one group, relying on the same strategies in previous studies (e.g., Aluja et al., 2019; Benko et al., 2010; Demetrovics et al., 2010; Goçi Uka et al., 2019; Light et al., 2007; Reuter et al., 2006; Smillie et al., 2010; Whitmer & Gotlib, 2012). There are at least two reasons for this decision. First, some alleles were extremely underrepresented, not meeting the basic prerequisites for statistical analyzes. Second, even in the case of genes that had a uniform distribution of alleles, we opted for this strategy due to the small sample size and the increase in the statistical power of the results. Genotype frequencies and allelic distributions are given in Table 1.
Data Analysis
Regarding the twin study, sample size estimation based on the RMSEA index (Kim, 2005) was conducted in the “semTools” package in R (Jorgensen et al., 2020; R Core Team, 2020). For the univariate AE model (two groups), with H0 RMSEA = .05, H1 RMSEA = .08, df = 4, alpha = .05, and power = .80, the required sample size is 628. With the H0 RMSEA lowered to .04 for the same model, the optimal sample size is 503, while for the H0 RMSEA of .03 the optimal sample size equals 398. Thus, although the sample can be considered to be smaller than required for the more complex models and smaller effects, apparently there is sufficient power to test the univariate AE models.
Genetic and environmental influences on phenotypic similarities between MZ and DZ were examined for each personality dimension by using structural equation modeling (SEM) – including univariate and multivariate biometric methods. Independent pathways (Fig. 1) and common pathways (Fig. 2) multivariate models were applied in order to estimate additive genetic factors (A); shared environmental (C), and non-shared environmental (E); and specific (s) and common (c) genetic and environmental sources of variance. In addition, Cholesky behavior genetic analyses (Loehlin, 1996) were calculated in order to explore genetic and environmental correlations among personality dimensions. Cholesky decomposition is often used in quantitative behavioral genetics to disentangle genetic and environmental influences (Neale & Maes, 2004.)
The overall model fit was estimated through several indices: the quotient χ2/df (optimal values tentatively mentioned as ranging from 2 to 3; Alavi et al., 2020; Cole, 1987; Kline, 2015); Comparative Fit Index and the Tucker-Lewis Index (CFI and TLI – optimal values higher than .95, acceptable higher than .90; Garson, 2015; Little, 2013), the Root Mean square Error of Approximation (RMSEA - optimal values lower than .05, acceptable lower than .08; Little, 2013), the Standardized Root Mean Square Residual (SRMR), with acceptable values below .08 (Brown, 2015), Akaike Information Criterion (AIC) and Bayesian information criterion (BIC), with lower values indicating better fit (Kline, 2015). The estimation method was maximum likelihood. Prior to the analyses, gender, and age (linear and quadratic) effects were controlled (partialized), while the lowest and highest 2% of scores were winsorized.
Multivariate SEM modeling was conducted in the “lavaan” package in R (Rosseel, 2012). R scripts for quantitative behavioral genetic analyses in the R environment are deposited at Zenodo (Čolović, 2019, https://doi.org/10.5281/zenodo.3514218).
Effect of specific gene polymorphisms were calculated using t-test for independent samples and Cohen’s d was calculated as a measure of effect size. A commonly used interpretation is to refer to effect sizes as small (d = .20), medium (d = .50), and large (d = .80), based on benchmarks suggested by Cohen (1992). rRST traits were dependent and allelic variants for COMT (rs4680), DRD2 (rs1800497), HTR1A (rs6295) and TPH2 (rs4570625) gene polymorphisms were independent variables.
Interactions/epistatic effects of selected gene polymorphisms on rRST personality dimensions were explored using univariate General linear models, computed with rRST traits as dependent and allelic variants of selected genes as the independent variables. Partial eta squared (ηp2) was used as a measure of effect size. A commonly used interpretation is to refer to effect sizes as small (ηp2 = .01), medium (ηp2 = .05), and large (ηp2 = .13), suggested by Cohen (1992).
All results are adjusted using Bonferroni correction for multiple comparisons. The analysis was carried out using the SPSS v.21 software (IBM corp., 2012). Power analyses for the interaction effects, as conceptualized by Lakens and Caldwell (2021), suggest that the power to correctly estimate approximately moderate main effects (ηp2 = .05) along with a large disordinal interaction (ηp2 = .15) in a 2 × 2 between-groups design, at alpha .01 and N = 350 approximates 100. The statistical power to capture a moderate (ηp2 = .06) effect in a t-test is over 99%, while a similar power estimation is obtained for a moderate effect in a one-way, three-group ANOVA design.
Results
Descriptive Statistics, Intra-class and Cross Twin – Cross Trait Correlations
Table 2 shows descriptive statistics for the used personality variables. Both types of phenotypic correlation coefficients (intraclass and cross twin – cross-trait) were calculated separately for monozygotic and dizygotic twins on rRST dimensions (Table 2).
Correlations between MZ twins are consistently higher than correlations between DZ twins on all variables. The largest intra-correlation difference was detected in BAS (Δr = .44), and the smallest in the case of Freeze (Δr = .13).
Univariate and Multivariate Genetic Modeling: Model Comparison and Parameter Estimation.
Although all proposed univariate biometric models, both full (ACE) and reduced (AE, CE) models, show good model fit, according to the BIC and AIC criteria, the univariate AE models fitted best for all rRST personality traits (Table 3). Fit indices were within acceptable boundaries for BIS, BAS, Fight, Flight and Freeze.
Furthermore, multivariate Independent pathway models and Common pathway models (the former comprising a single latent factor) were tested, regarding the specification of the form of the observed phenotypic correlations among rRST personality traits. A comparison of the two sets of models, as well as the comparison between full (ACE) and reduced (AE, CE) models, was carried out using several fit indices.
Fit indices for the common pathways AE model (Table 3) are the most appropriate and within acceptable boundaries, χ2/df = 1.40, CFI = .92, TLI = .92, RMSEA = .06, AIC = 10,337.48, BIC = 10,461.32, except SRMR, which is above .08 (SRMR = .09). The estimation of the parameters of the common AE model is given in Table 3.
It is noticeable in Table 4 that all personality traits have moderate to strong genetic bases (from 37% for Flight to 44% for Freeze). Also, common genetic influences range from 37% (for Freeze) to 0.4% (for Fight). In contrast, unique genetic factors are most prominent in Fight (35%), BAS (32%), and BIS (18%). Common environmental impacts are generally low, with the exception for BIS (50%) and Freeze (17%). Moreover, unique environmental contributions are moderate to strong (from 61% for Fight, Flight, and BAS - 57% and 52%, to 39% for Freeze).
Cholesky behavior genetic analyses are performed to explore genetic and environmental correlations of the rRST personality dimensions (Table 5). The strongest genetic correlation is observed between Freeze and BIS (.70), Freeze and Flight (.71) and Flight and BIS (.56), as well as BAS and Flight (−.59). The weakest genetic correlation is between Freeze and Fight (−.17). Environmental correlations are weak to moderate; the strongest correlation is .48 (BIS and Freeze), then BIS and BAS (−.29), and Flight and Freeze (.29). The weakest environmental correlation is between BIS and Fight (.16).
Allelic effects of COMT, DRD2, HTR1A and TPH2 polymorphisms on rRST dimensions.
The mean scores of all rRST variables in the specific allelic configurations of COMT, DRD2, TPH2 and HTR1A are given in Table 6.
There are two significant differences (effects) of genes on the rRTS dimensions (Table 7). It is the main effect of the COMT gene on the BAS (t = −2.32; p = .02, after Bonferroni p = .04; d = .24, suggesting small to medium effect size). Participants with Val genotype were scoring higher on BAS. The other effect is the TPH2 gene on the BIS (t = 2.79; p = .01, after Bonferroni p = .02, d = .29, suggesting small to medium effect size). Participants with dominant TPH2 GG genotype have significantly higher scores on BIS than carriers with TPH2 GT genotype.
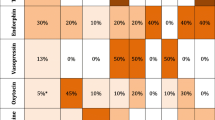
Results of epistatic genetic effects on rRST dimensions are given in Table 8. The interaction effect of COMT and DRD2 genotypes on BAS (F = 9.64; p = .001; ηp2 = .02, suggesting small effect size) was significant before and after correction for multiple comparison (after Bonferroni correction, adjusted p level was still significant, p = .004). Participants with COMT Met+ genotype and DRD2 A1+ genotype had the lowest score on BAS. The direction of the interaction effect is shown in Fig. 3.
There was also a significant interaction effect of HTR1A and TPH2 genotypes on Fight (F = 13.59; p = .001; ηp2 = .03, suggesting small effect size). After Bonferroni correction, the effect remained significant and robust (p = .004). Participants with HTR1A CC genotype and TPH2 GG genotype had the lowest score on Fight (Table 8). The direction of the interaction effect is shown in Fig. 4.
Discussion
A fundamental finding from behavior genetic research is that personality traits are heritable (Plomin et al., 2016). However, the problems in the conceptualization of personality constructs affect how we examine the heritability of new constructs, resulting from recent insights into the neurophysiological basis of behavior (Gray & McNaughton, 2000). Although the rRST becomes the dominant paradigm for the biological understanding of neuropsychological functioning (Corr, 2016), this is the first behavioral genetic study based on reformulated dimensions of BIS, BAS and FFFS dimensions.
The pattern of findings provides the first behavioral genetic support for the assertion that variables measured by the RSQ are adequate phenotypic descriptions of reward and punishment reactions defined by the Reinforcement sensitivity theory (Gray & McNaughton, 2000). The heritability estimates for all dimensions range from .34 to .44. Genetic correlation patterns indicate a clear distinction between approaching and avoiding responses. Namely, there are significant genetic and environmental correlations between BIS, Flight and Freeze systems, which points to the possible general hereditary basis of avoiding behavior, as well as a similar pattern of learning and adopting avoidance behavior in coping with dangers. Although the rRST makes a clear distinction between responses to potential and real danger (Gray & McNaughton, 2000), it is possible that there are similar genetic mechanisms which underlie all types of avoidant behavior. It is also possible that the learned avoidance reaction is generalized to all perceived threatening situations. Moreover, there are significant negative correlations between these dimensions and BAS and Fight, which points to the possibility that the avoidance behavior is opposed to approach, which creates an “or-or” schema reactivity to reward and punishment. In other words, there seems to be a strong genetic basis for responsiveness to reward and punishment, which shapes the development of one of two possible strategies – approach or avoidance. Furthermore, the genetic and environmental correlations between BAS and Fight are significant, which is a sufficient argument for the view that there are some similar genetic and environmental factors in the basis of the approaching reactions. These results support the approach-avoidance hypothesis (Corr & McNaughton, 2012), which explains the phenotypic similarities between the reactions to appetitive and aversive stimuli from the environment.
However, the multivariate model confirms the existence of only one factor, determined by the additive genetic and nonshared environmental variance. It points to the existence of a common genetic factor for BIS, Fight and Freeze, while BAS and Fight do not load on the common factor, which would be in favor of the existence of a specific approach-related factor. Instead, both BAS and Fight are explained by specific genetic factors and unique nonshared environmental factors. This result points to the possibility that different types of approach behavior have different genetic bases. It is possible that aggression (Fight) has various behavioral outcomes, which do not necessarily involve lack of control, which is characteristic of impulsivity (BAS). Similarly, impulsive behavior does not necessarily involve aggression. Although the “avoiding” dimensions share a common genetic variance, there is a substantial proportion of unique genetic variance, characteristic for BIS and Flight, and the least likely for Freeze. Also, the common nonshared environmental variance is most pronounced in BIS. Nevertheless, although the environmental factors shape the specific learning responses to threatening stimuli, the results indicate the existence of similarities in the genetic basis of the avoiding reactions, such as BIS, Flight, and Freeze.
Molecular genetic analyses on the same twin sample indicate that the dimensions of the rRST are defined by different gene underlying specific responses, which is a key argument for the independent phenotypic description. The main effect of TPH2 on BIS indicates the association of GG homozygotes with higher scores on anxiety. This result is consistent with results that confirm the relationship between G allele and anxiety or neuroticism (Laasa et al., 2017). Namely, previous results showed that the neuroticism score intended to be lower in the TPH2 TT homozygotes (Laasa et al., 2017) and that the correlation between amygdala – hippocampus connectivity and RST anxiety is positive in individuals with the TPH2 G allele (Hahn et al., 2013). Despite its role in the regulation of anxiety, the TPH2 gene has an important contribution to the regulation of aggressive behavior. Our results demonstrated that interaction of the HTR1A and TPH2 has a significant effect on the Fight. Specifically, the lowest score on Fight have participants with HTR1A CC and TPH2 GG genotypes. This result indicates the epistatic effect of serotonin-regulating genes in aggression. Although previous research has highlighted the role of TPH2 gene (Waider et al., 2011) and HTR1A (Witte et al., 2009) in aggression, their specific combination apparently leads to reduced level of serotonin, which contributes to a lack of affective regulation.
Although the main effect of COMT on BAS is expected, the direction of association is not consistent with the dominant previous findings (Chen et al., 2011; Demetrovics et al., 2010; Hoth et al., 2006). Namely, Met alleles are commonly associated with high impulsivity, which makes our result contradictory. Since this effect is not robust, it is possible that it is an artifact due to the size of the sample. However, the results of our study confirm the previous findings (Reuter et al., 2006) that the interaction between COMT and DRD2 gene polymorphisms has a significant effect on BAS. Reuter et al. (2006) found a significant association with two of the three BAS subscales from the BIS/BAS scale (Carver & White, 1994), which evaluate behavioral approach tendencies from a slightly different perspective than the RSQ (Smederevac et al., 2014). Nevertheless, the impulsivity within the BAS system can be at least partly explained by the epistatic effect of the COMT and DRD2 gene. Although there was no main effect observed for each of these gene loci, we identified an association between the interaction of the Met+/A1+ with BAS. Other genotype combinations (i.e., Met+/A1-, Met−/A1+ and Met−/A1-) did not exhibit significant relations to BAS score. Our results may partially confirm previous findings (Reuter et al., 2006) that equilibrium between catabolic enzyme activity of COMT and DRD2 receptor density is associated with dopamine levels and predicts impulsivity. More specifically, the high level of dopamine is regulated by a larger number of genes and is associated with a lower score on BAS. Inversely, this result would suggest that higher reward sensitivity and stronger approach behavior may be related to a structural deficit in the dopaminergic reward system.
In our sample, DRD2, COMT, HTR1A and TPH2 genes were not significantly related to Flight and Freeze scores. This result points to a different molecular genetic basis of anxiety and fear, as one of the basic arguments for the existence of specific systems within rRST (Gray & McNaughton, 2000). Anxiety is regulated by the TPH2 gene, which influences serotonin levels. Despite phenotypic similarities between these dimensions and similarities in heritability patterns, TPH2 gene is linked only to BIS, but not to Flight and Freeze. Although it could be expected that the molecular genetic basis of the so-called “avoiding” behaviors will be similar, this result highlights the differences, which need to be further examined, considering a larger number of genes.
Moreover, the differences in the molecular genetic basis of the so-called “approach” behavior also differ, since the interaction of the DRD2 and the COMT genes is related to BAS, while the interaction of the HTR1A and TPH2 are associated with high scores on Fight. These results indicate that serotonin levels are dependent on the epistatic effect of multiple genes. Fight is defined as one of the possible reactions to the real threat that provokes emotion of fear. It is possible that the emotional aspect of the aggressive reaction is regulated by genes such as HTR1A and TPH2, indicating that the aggressive response may appear only in conditions of increased tension.
These results need to be interpreted with great caution, since there is an increased likelihood for false positive findings in this type of twin and candidate gene studies due to small sample size. Regarding genetic association study, the sample size is clearly underpowered. Therefore, only robust effects were considered, which underwent Bonferroni correction.
Although the twin sample is valuable for behavioral genetic studies, it is still rare in molecular genetic analyses. Nevertheless, these results represent the robust confirmation of previous findings on non-twin samples (Hahn et al., 2013; Kautzky et al., 2019; Laasa et al., 2017; Reuter et al., 2006), which is important for the process of validating the scientific results which associate specific genes with complex behaviors. An important limitation of the study may be related to the number of gene polymorphisms analyzed. We are aware that in the era of Genome Wide Associations Studies (GWAS) (Montag et al., 2020), four selected genes cover a small part of variability of the genetic base of behavior. However, the selection of those genes that are often linked to basic personality traits facilitates the potential replication of these specific findings.
Conlusions
This study provides the first quantitative behavioral genetic evidence on the genetic basis of approach-avoidance strategies using rRST background (Corr & McNaughton, 2012). Namely, there are clear genetic and environmental correlations between the approaching dimensions BAS and Fight, which correspond to impulsivity and aggressiveness, as well as between the avoiding dimensions BIS, Flight, and Freeze, which correspond to anxiety, fear, and panic. However, the essential specificities in the molecular-genetic basis of these dimensions represent a demarcation line of individual differences within the general responses to reward and punishment. Namely, avoidance dimensions BIS, Flight and Freeze share common genetic variance, with a significant portion of the unique genetic variance. The association of the TPH2 only with BIS indicates a clear genetic difference between anxiety and fear. In other words, within the general avoidance strategy, there are specific genetic patterns that allow the distinction between anxiety, fear, and panic. Also, the approaching behaviors, such as BAS and Fight share a part of the common genetic variance, while each dimension has a unique genetic variance, which was illustrated by molecular genetic analyzes. The interaction of dopamine genes in the prediction of BAS and serotonin genes in the prediction of Fight indicates substantial genetic differences between impulsivity and aggressiveness. Within the general approach strategy, there are specific genetic patterns that allow the distinction between impulsivity and aggressiveness.
Our results indicate a large contribution of environmental factors in shaping the genetic readiness to recognize and respond to various appetitive and aversive stimuli. The finding that unique environmental contributions to dimensions range from 39% to 61% points to the fact that learning processes and previous experience are key to adopting specific patterns of adaptation.
Finally, although the simultaneous application of the quantitative behavioral genetic and molecular genetic approaches cannot warrant the complete agreement among their respective results, combining these two methodological approaches on the same sample of twins enabled the future reproducibility of the findings, especially those related to common and specific genetic and environmental contributions to approach and avoidance strategies.
Data Availability
The datasets generated and analysed during the current study are available in the OSF repository, [https://osf.io/csh23/].
References
Aarts, E., Roelofs, A., Franke, B., Rijpkema, M., Fernández, G., Helmich, R. C., & Cools, R. (2010). Striatal Dopamine Mediates the Interface between Motivational and Cognitive Control in Humans: Evidence from Genetic Imaging. Neuropsychopharmacology, 35(9), 1943–1951. https://doi.org/10.1038/npp.2010.68.
Akil, M., Kolachana, B. S., Rothmond, D. A., Hyde, T. M., Weinberger, D. R., & Kleinman, J. E. (2003). Catechol-o-methyltransferase genotype and dopamine regulation in the human brain. The Journal of Neuroscience, 23, 2008–2013. https://doi.org/10.1523/jneurosci.23-06-02008.2003
Alavi, M., Visentin, D. C., Thapa, D. K., Hunt, G. E., Watson, R., & Cleary, M. (2020). Chi-square for model fit in confirmatory factor analysis. Journal of Advanced Nursing, 76(9), 2209–2211. https://doi.org/10.1111/jan.14399
Alqahtani, A. M., Kumarappan, C., Kumar, V., Srinivasan, R., & Krishnaraju, V. (2020). Understanding the genetic aspects of resistance to antidepressants treatment. European Review for Medical and Pharmacological. Science., 24, 7784–7795. https://doi.org/10.26355/eurrev_202007_22281
Aluja, A., Balada, F., Blanco, E., Fibla, J., & Blanch, A. (2019). Twenty candidate genes predicting neuroticism and sensation seeking personality traits: A multivariate analysis association approach. Personality and Individual Differences, 140, 90–102. https://doi.org/10.1016/j.paid.2018.03.041
Aoki, J., Ikeda, K., Murayama, O., Yoshihara, E., Ogai, Y., & Iwahashi, K. (2010). The association between personality, pain threshold and a single nucleotide polymorphism (rs3813034) in the 3′-untranslated region of the serotonin transporter gene (SLC6A4). Journal of Clinical Neuroscience, 17, 574–578. https://doi.org/10.1016/j.jocn.2009.08.020
Benko, A., Lazary, J., Molnar, E., Gonda, X., Tothfalusi, L., Pap, D., et al. (2010). Significant association between the C (− 1019) G functional polymorphism of the HTR1A gene and impulsivity. American Journal of Medical Genetics Part B: Neuropsychiatric Genetics, 153, 592–599. https://doi.org/10.1002/ajmg.b.31025
Bezdjian, S., Baker, L. A., & Tuvblad, C. (2011). Genetic and environmental influences on impulsivity: A meta-analysis of twin, family and adoption studies. Clinical Psychology Review, 31, 1209–1223. https://doi.org/10.1016/j.cpr.2011.07.005
Bienvenu, O. J., Davydow, D. S., & Kendler, K. S. (2011). Psychiatric ‘diseases’ versus behavioral disorders and degree of genetic influence. Psychological Medicine, 41, 33–40. https://doi.org/10.1017/s003329171000084x
Brown, T. A. (2015). Confirmatory factor analysis for applied research (2nd ed. (pp. xvii, 462). The Guilford Press.
Carver, C. S., & White, T. L. (1994). Behavioral inhibition, behavioral activation, and affective responses to impending reward and punishment: The BIS/BAS Scales. Journal of Personality and Social Psychology, 67, 319–333. https://doi.org/10.1037//0022-3514.67.2.319
Chen, C., Chen, C., Moyzis, R., Dong, Q., He, Q., Zhu, B., et al. (2011). Sex modulates the associations between the COMT gene and personality traits. Neuropsychopharmacology, 36, 1593–1598. https://doi.org/10.1038/npp.2011.39
Coccaro, E. F., Bergeman, C., & McClearn, G. E. (1993). Heritability of irritable impulsiveness: A study of twins reared together and apart. Psychiatry Research, 48, 229–242. https://doi.org/10.1016/0165-1781(93)90074-q
Cohen, J. (1992). A power primer. Psychological Bulletin, 112, 155–159. https://doi.org/10.1037/0033-2909.112.1.155
Cole, D. A. (1987). Utility of confirmatory factor analysis in test validation research. Journal of consulting and clinical psychology, 55(4), 584.
Čolović, P. (2019). petarcolovic. Petarcolovic/R_CqBG 1.0.0-Beta. Zenodo, 2019. Zenodo. https://doi.org/10.5281/zenodo.3514218
Corr, P. J. (2016). Reinforcement sensitivity theory of personality questionnaires: Structural survey with recommendations. Personality and Individual Differences, 89, 60–64. https://doi.org/10.1016/j.paid.2015.09.0450191-8869
Corr, P. J., & Cooper, A. (2016). The Reinforcement Sensitivity Theory of Personality Questionnaire (RST-PQ): Development and validation. Psychological Assessment, 28, 1427–1440. https://doi.org/10.1037/pas0000273
Corr, P. J., & McNaughton, N. (2012). Neuroscience and approach/avoidance personality traits: A two stage (valuation-motivation) approach. Neuroscience & Biobehavioral Reviews, 36, 2239–2354. https://doi.org/10.1016/j.neubiorev.2012.09.013
Demetrovics, Z., Varga, G., Szekely, A., Vereczkei, A., Csorba, J., Balazs, H., et al. (2010). Association between Novelty Seeking of opiate-dependent patients and the catechol-O-methyltransferase Val158Met polymorphism. Comprehensive Psychiatry, 51, 510–515. https://doi.org/10.1016/j.comppsych.2009.11.008
DeYoung, C. G., Cicchetti, D., Rogosch, F. A., Gray, J. R., Eastman, M., & Grigorenko, E. L. (2011). Sources of cognitive exploration: Genetic variation in the prefrontal dopamine system predicts Openness/Intellect. Journal of research in personality, 45, 364–371. https://doi.org/10.1016/j.jrp.2011.04.002
Gagne, J. R., Osullivan, D. L., Schmidt, N. L., Spann, C. A., & Goldsmith, H. H. (2016). The shared etiology of attentional control and anxiety: An adolescent twin study. Journal of Research on Adolescence, 27, 122–138. https://doi.org/10.1111/jora.12260
Garcia-Garcia, M., Barceló, F., Clemente, I., & Escera, C. (2011). COMT and ANKK1 gene–gene interaction modulates contextual updating of mental representations. NeuroImage, 56, 1641–1647. https://doi.org/10.1016/j.neuroimage.2011.02.053
Garson, G. D. (2015). Structural Equation Modeling. Statistical Associates Publishers.
Goçi Uka, A., Agani, F., Blyta, A., Hoxha, B., Haxhibeqiri, S., Haxhibeqiri, V., et al. (2019). Role of the allelic variation in the 5-hydroxytryptamine receptor 1A (HTR1A) and the tryptophan hydroxylase 2 (TPH2) genes in the development of PTSD. Psychiatria Danubina, 31(2), 256–262. https://doi.org/10.24869/psyd.2019.256
Goldman, D., Oroszi, G., & Ducci, F. (2005). The genetics of addictions: Uncovering the genes. Nature Reviews Genetics, 6, 521–532. https://doi.org/10.1038/nrg1635
Gray, J. A. (1982). The neuropsychology of anxiety: An enquiry into the functions of the septo-hippocampal system (1st ed.). Oxford University Press.
Gray, J. A., & McNaughton, N. (2000). The neuropsychology of anxiety: An enquiry into the functions of the septo-hippocampal system (2nd ed.). Oxford University Press.
Gray, J. C., MacKillop, J., Weafer, J., Hernandez, K. M., Gao, J., Palmer, A. A., & de Wit, H. (2018). Genetic analysis of impulsive personality traits: Examination of a priori candidates and genome-wide variation. Psychiatry Research, 259, 398–404. https://doi.org/10.1016/j.psychres.2017.10.047
Gutknecht, L., Jacob, C., Strobel, A., Kriegebaum, C., Müller, J., Zeng, Y., et al. (2007). Tryptophan hydroxylase-2 gene variation influences personality traits and disorders related to emotional dysregulation. International Journal of Neuropsychopharmacology, 10, 309–320. https://doi.org/10.1017/S1461145706007437
Hahn, T., Heinzel, S., Notebaert, K., Dresler, T., Reif, A., Lesch, K.-P., et al. (2013). The tricks of the trait: Neural implementation of personality varies with genotype-dependent serotonin levels. NeuroImage, 81, 393–399. https://doi.org/10.1016/j.neuroimage.2013.05.037
Hettema, J. M., Annas, P., Neale, M. C., Kendler, K. S., & Fredrikson, M. (2003). A twin study of the genetics of fear conditioning. Archives of General Psychiatry, 60, 702. https://doi.org/10.1001/archpsyc.60.7.702
Hoth, K. F., Paul, R. H., Williams, L. M., Dobson-Stone, C., Todd, E., Schofield, P. R., et al. (2006). Associations between the COMT Val/Met polymorphism, early life stress, and personality among healthy adults. Neuropsychiatric Disease and Treatment, 2, 219. https://doi.org/10.2147/nedt.2006.2.2.219
Houtem, C. V., Laine, M., Boomsma, D., Ligthart, L., Wijk, A. V., & Jongh, A. D. (2013). A review and meta-analysis of the heritability of specific phobia subtypes and corresponding fears. Journal of Anxiety Disorders, 27, 379–388. https://doi.org/10.1016/j.janxdis.2013.04.007
IBM corporation (2012). IBM SPSS Statistics for Windows, version 21.0. IBM Corp.
Jackson, C. J. (2009). Jackson-5 scales of revised Reinforcement Sensitivity Theory (r-RST) and their application to dysfunctional real world outcomes. Journal of Research in Personality, 43, 556–569. https://doi.org/10.1016/j.jrp.2009.02.007
Johnson, S. L., Carver, C. S., Joormann, J., & Cuccaro, M. L. (2016). Genetic polymorphisms related to behavioral approach and behavioral inhibition scales. Personality and Individual Differences, 88, 251–255. https://doi.org/10.1016/j.paid.2015.09.024
Jorgensen, T. D., Pornprasertmanit, S., Schoemann, A. M., & Rosseel, Y. (2020). semTools: Useful tools for structural equation modeling. R package version 0.5–3. Retrieved from https://CRAN.R-project.org/package=semTools at May, 2021.
Juhasz, G., Downey, D., Hinvest, N., Thomas, E., Chase, D., Toth, Z. G., Lloyd-Williams, K., Mekli, K., Platt, H., Payton, A., Bagdy, G., Elliott, R., Deakin, J. F. W., & Anderson, I. M. (2009). Risk-Taking Behavior in a Gambling Task Associated with Variations in the Tryptophan Hydroxylase 2 Gene: Relevance to Psychiatric Disorders. Neuropsychopharmacology, 35(5), 1109–1119. https://doi.org/10.1038/npp.2009.216
Kalbitzer, J., Frokjaer, V. G., Erritzoe, D., Svarer, C., Cumming, P., Nielsen, F. Å., et al. (2009). The personality trait openness is related to cerebral 5-HTT levels. Neuroimage, 45, 280–285. https://doi.org/10.1016/j.neuroimage.2008.12.001
Kautzky, A., James, G. M., Philippe, C., Baldinger-Melich, P., Kraus, C., Kranz, G. S., et al. (2019). Epistasis of HTR1A and BDNF risk genes alters cortical 5-HT1A receptor binding: PET results link genotype to molecular phenotype in depression. Translational Psychiatry, 9. https://doi.org/10.1038/s41398-018-0308-2
Kim, K. H. (2005). The relation among fit indexes, power, and sample size in structural equation modeling. Structural Equation Modeling, 12, 368–390.
Kline, R. B. (2015). Principles and Practice of Structural Equation Modeling, Fourth Edition (4th ed.). The Guilford Press.
Koller, G., Bondy, B., Preuss, U. W., Zill, P., & Soyka, M. (2006). The C (−1019) G 5-HT1A promoter polymorphism and personality traits: no evidence for significant association in alcoholic patients. Behavioral and Brain Functions, 2, 7. https://doi.org/10.1186/1744-9081-2-7
Kotyuk, E., Duchek, J., Head, D., Szekely, A., Goate, A. M., & Balota, D. A. (2015). A genetic variant (COMT) coding dopaminergic activity predicts personality traits in healthy elderly. Personality and Individual Differences, 82, 61–66. https://doi.org/10.1016/j.paid.2015.03.012
Laasa, K., Kiiveb, E., Mäestuc, J., Vahta, M., Veidebaumd, T., & Harro, J. (2017). Nice guys: Homozygocity for the TPH2 -703G/T (rs4570625) minor allele. Journal of Affective Disorders, 215, 230–236. https://doi.org/10.1016/j.jad.2017.03.045
Lakens, D., & Caldwell, A. (2021). Simulation-Based Power Analysis for Factorial Analysis of Variance Designs. Advances in Methods and Practices in Psychological Science, 4(1), 251524592095150. https://doi.org/10.1177/2515245920951503
Lehto, K., Vaht, M., Mäestu, J., Veidebaum, T., & Harro, J. (2015). Effect of tryptophan hydroxylase-2 gene polymorphism G-703 T on personality in a population representative sample. Progress in Neuro-Psychopharmacology and Biological Psychiatry, 57, 31–35. https://doi.org/10.1016/j.pnpbp.2014.10.005
Lemonde, S., Turecki, G., Bakish, D., et al. (2003). Impaired repression at a 5- hydroxytryptamine 1A receptor gene polymorphism associated with major depression and suicide. Journal of Neuroscience, 23, 8788–8799. https://doi.org/10.1523/jneurosci.23-25-08788.2003
Light, K. J., Joyce, P. R., Luty, S. E., Mulder, R. T., Carter, J. D., Frampton, C. M., Miller, A. L., & Kennedy, M. A. (2007). An association study of DRD2 and COMT polymorphisms with novelty seeking and harm avoidance scores, in two independent samples of depressed patients. Behavioral and Brain Functions, 3, 3. https://doi.org/10.1186/1744-9081-3-3
Little, T. D. (2013). Longitudinal Structural Equation Modeling. Guilford Press.
Loehlin, J. C. (1996). The Cholesky approach: A cautionary note. Behavior Genetics, 26, 65–69. https://doi.org/10.1007/BF02361160
Mandelli, L., Antypa, N., Nearchou, F. A., Vaiopoulos, C., Stefanis, C. N., Serretti, A., & Stefanis, N. C. (2012). The role of serotonergic genes and environmental stress on the development of depressive symptoms and neuroticism. Journal of Affective Disorders, 142(1–3), 82–89. https://doi.org/10.1016/j.jad.2012.03.047
Mier, D., Kirsch, P., & Meyer-Lindenberg, A. (2010). Neural substrates of pleiotropic action of genetic variation in COMT: a meta-analysis. Molecular Psychiatry, 15, 918–927. https://doi.org/10.1038/mp.2009.36
Mihić, L., Čolović, P., Ignjatović, I., Smederevac, S., & Novović, Z. (2015). Anxiety between personality and cognition: The gray zone. Personality and Individual Differences, 78, 19–23. https://doi.org/10.1016/j.paid.2015.01.013.
Mitrović, D., Smederevac, S., Čolović, P., Kodžopeljić, J., & Dinić, B. (2014). Personality prototypes based on dimensions of the revised reinforcement sensitivity theory among prisoners and non prisoners. Personality and Individual Differences, 69, 50–55. https://doi.org/10.1016/j.paid.2014.05.004
Montag, C., Ebstein, R. P., Jawinski, P., & Markett, S. (2020). Molecular genetics in psychology and personality neuroscience: On candidate genes, genome wide scans, and new research strategies. Neuroscience & Biobehavioral Reviews, 118, 163–174. https://doi.org/10.1016/j.neubiorev.2020.06.020
Montag, C., Markett, S., Basten, U., Stelzel, C., Fiebach, C., Canli, T., & Reuter, M. (2010). Epistasis of the DRD2/ANKK1 Taq Ia and the BDNF Val66Met polymorphism impacts novelty seeking and harm avoidance. Neuropsychopharmacology, 35, 1860–1867. https://doi.org/10.1038/npp.2010.55
Neale, M. C., & Maes, H. H. M. (2004). Methodology for Genetic Studies of Twins and Families. Kluwer Retrieved from https://ibgwww.colorado.edu/workshop2004/cdrom/HTML/book2004a.pdf
Nelson, R. J., & Trainor, B. C. (2007). Neural mechanisms of aggression. Nature Reviews Neuroscience, 8, 536–546. https://doi.org/10.1038/nrn2174
Pearson, R., McGeary, J. E., & Beevers, C. G. (2014). Association between serotonin Cumulative Genetic Score and the Behavioral Approach System (BAS): Moderation by early life environment. Personality and Individual Differences, 70, 140–144. https://doi.org/10.1016/j.paid.2014.06.041
Pecina, M., Mickey, B. J., Love, T., Wang, H., Langenecker, S. A., Hodgkinson, C., et al. (2013). DRD2 polymorphisms modulate reward and emotion processing, dopamine neurotransmission and openness to experience. Cortex, 49, 877–890. https://doi.org/10.1016/j.cortex.2012.01.010
Plomin, R., Defries, J. C., Knopik, V. S., & Neiderhiser, J. M. (2016). Top 10 replicated findings from behavioral genetics. Perspectives on Psychological Science, 11, 3–23. https://doi.org/10.1177/1745691615617439
R Core Team. (2020). R: A language and environment for statistical computing. R Foundation for Statistical Computing. https://www.R-project.org/
Ranđelović, K., Smederevac, S., Čolović, P., & Corr, P. (2018). Fear and anxiety in social setting: an experimental study. Journal of Individual Differences, 39, 61–75. https://doi.org/10.1027/1614-0001/a000251
Reuter, M., Cooper, A. J., Smillie, L. D., Markett, S., & Montag, C. (2015). A new measure for the revised reinforcement sensitivity theory: Psychometric criteria and genetic validation. Frontiers in Systems Neuroscience, 9. https://doi.org/10.3389/fnsys.2015.00038
Reuter, M., Schmitz, A., Corr, P., & Hennig, J. (2006). Molecular genetics support Gray’s personality theory: The interaction of COMT and DRD2 polymorphisms predicts the behavioural approach system. International Journal of Neuropsychopharmacology, 9, 155–166. https://doi.org/10.1017/s1461145705005419
Rosseel, Y. (2012). Lavaan: An R package for structural equation modeling. Journal of Statistical Software, 48, 1–36 Retrieved from http://www.jstatsoft.org/v48/i02/
Sadiković, S., Branovački, B., Oljača, M., Mitrović, D., Pajić, D., & Smederevac, S. (2020). Daily Monitoring of Emotional Responses to the Coronavirus Pandemic in Serbia: A Citizen Science Approach. Frontiers in Psychology, 11. https://doi.org/10.3389/fpsyg.2020.02133
Schinka, J. A., Busch, R. M., & Robichaux-Keene, N. (2004). A meta-analysis of the association between the serotonin transporter gene polymorphism (5-HTTLPR) and trait anxiety. Molecular Psychiatry, 9, 197–202. https://doi.org/10.1038/sj.mp.4001405
Smederevac, S., Mitrović, D., Čolović, P., & Nikolašević, Ž. (2014). Validation of the measure of revised Reinforcement Sensitivity Theory constructs. Journal of Individual Differences, 35, 12–21. https://doi.org/10.1027/1614-0001/a000121
Smederevac, S., Mitrović, D., Sadiković, S., et al. (2019). Serbian Twin Registry. Twin Research and Human Genetics, 22, 660–666. https://doi.org/10.1017/thg.2019.114
Smillie, L. D., Cooper, A. J., Proitsi, P., Powell, J. F., & Pickering, A. D. (2010). Variation in DRD2 dopamine gene predicts extraverted personality. Neuroscience Letters, 468, 234–237. https://doi.org/10.1016/j.neulet.2009.10.095
Spagnolo, P. A., Norato, G., Maurer, C. W., Goldman, D., Hodgkinson, C., Horovitz, S., & Hallett, M. (2020). Effects of TPH2 gene variation and childhood trauma on the clinical and circuit-level phenotype of functional movement disorders. Journal of Neurology, Neurosurgery & Psychiatry, 91(8), 814–821. https://doi.org/10.1136/jnnp-2019-322636
Strobel, A., Gutknecht, L., Zheng, Y., Reif, A., Brocke, B., & Lesch, K. P. (2003). Allelic variation of serotonin receptor 1A function is associated with anxiety- and depression-related traits. Journal of Neural Transmission, 110, 1445–1453. https://doi.org/10.1007/s00702-003-0072-0
Takahashi, Y., Yamagata, S., Kijima, N., Shigemasu, K., Ono, Y., & Ando, J. (2007). Continuity and change in behavioral inhibition and activation systems: A longitudinal behavioral genetic study. Personality and Individual Differences, 43, 1616–1625. https://doi.org/10.1016/j.paid.2007.04.030
Tsuchimine, S., Yasui-Furukori, N., Sasaki, K., Kaneda, A., Sugawara, N., Yoshida, S., & Kaneko, S. (2012). Association between the dopamine D2 receptor (DRD2) polymorphism and the personality traits of healthy Japanese participants. Progress in Neuro-Psychopharmacology and Biological Psychiatry, 38, 190–193. https://doi.org/10.1016/j.pnpbp.2012.03.008
Tunbridge, E. M., Narajos, M., Harrison, C. H., Beresford, C., Cipriani, A., & Harrison, P. J. (2019). Which dopamine polymorphisms are functional? systematic review and meta-analysis of COMT, DAT, DBH, DDC, DRD1–5, MAOA, MAOB, TH, VMAT1, and VMAT2. Biological Psychiatry, 86, 608–620. https://doi.org/10.1016/j.biopsych.2019.05.014
Valeeva, E. V., Kashevarov, G. S., Kasimova, R. R., Ahmetov, I. I., & Kravtsova, O. A. (2020). Association of the Val158Met Polymorphism of the COMT Gene with Measures of Psychophysiological Status in Athletes. Neuroscience and Behavioral Physiology, 50. https://doi.org/10.1007/s11055-020-00924-z
Waider, J., Araragi, N., Gutknecht, L., & Lesch, K.-P. (2011). Tryptophan hydroxylase-2 (TPH2) in disorders of cognitive control and emotion regulation: A perspective. Psychoneuroendocrinology, 36, 393–405. https://doi.org/10.1016/j.psyneuen.2010.12.012
Whisman, M. A., Richardson, E. D., & Smolen, A. (2011). Behavioral inhibition and triallelic genotyping of the serotonin transporter promoter (5-HTTLPR) polymorphism. Journal of Research in Personality, 45, 706–709. https://doi.org/10.1016/j.jrp.2011.08.009
Whitmer, A. J., & Gotlib, I. H. (2012). Depressive rumination and the C957T polymorphism of the DRD2 gene. Cognitive, Affective, & Behavioral Neuroscience, 12, 741–747. https://doi.org/10.3758/s13415-012-0112-z
Witte, A. V., Flöel, A., Stein, P., Savli, M., Mien, L.-K., Wadsak, W., et al. (2009). Aggression is related to frontal serotonin-1A receptor distribution as revealed by PET in healthy subjects. Human Brain Mapping, 30, 2558–2570. https://doi.org/10.1002/hbm.20
Code Availability
R scripts for quantitative behavioral genetic analyses in R environment are deposited at Zenodo (Čolović, 2019, https://doi.org/10.5281/zenodo.3514218).
Funding
This research was supported by the Science Fund of the Republic of Serbia (#7744418 GENIUS) and the German Research Foundation (DFG) (#220286500).
Author information
Authors and Affiliations
Contributions
Snežana Smederevac: Conceptualization, Methodology, Investigation, Writing - Original Draft, Funding acquisition, Selka Sadiković: Investigation, Writing - Original Draft, Data Curation, Petar Čolović: Methodology, Software, Formal analysis, Data Curation, Visualization, Nataša Vučinić: Investigation, Writing - Original Draft, Data Curation, Aleksandra Milutinović: Investigation, Writing - Original Draft, Data Curation, Rainer Riemann: Supervision, Writing - Review & Editing, Funding acquisition, Philip J Corr: Supervision, Writing - Review & Editing, Mechthild Prinz: Resources, Investigation, Supervision, Writing - Review & Editing, Zoran Budimlija: Resources, Supervision, Writing - Review & Editing.
Corresponding author
Ethics declarations
Ethics Approval
The research was approved by the Ethics Committee of the Faculty of Philosophy (#02-374/15) and the Committee for Ethics of Clinical Trials at the Faculty of Medicine (#01-39/229/1) at the University of Novi Sad.
Consent to Participate
Informed consent was obtained from all individual participants included in the study.
Consent for Publication
Not applicable.
Conflicts of Interest/Competing Interests
The authors have no relevant financial or non-financial interests to disclose.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Smederevac, S., Sadiković, S., Čolović, P. et al. Quantitative behavioral genetic and molecular genetic foundations of the approach and avoidance strategies. Curr Psychol 42, 14268–14282 (2023). https://doi.org/10.1007/s12144-022-02724-9
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12144-022-02724-9